6K7N
 
 | | Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E1P state) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Cell cycle control protein 50A, ... | | 著者 | Hiraizumi, M, Yamashita, K, Nishizawa, T, Nureki, O. | | 登録日 | 2019-06-07 | | 公開日 | 2019-08-28 | | 最終更新日 | 2021-02-10 | | 実験手法 | ELECTRON MICROSCOPY (2.84 Å) | | 主引用文献 | Cryo-EM structures capture the transport cycle of the P4-ATPase flippase.
Science, 365, 2019
|
|
5RS7
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000034618676 | | 分子名称: | 1-{2-[(propan-2-yl)oxy]ethyl}-2-sulfanylidene-1,2,3,5-tetrahydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
6VAJ
 
 | | Crystal Structure Analysis of human PIN1 | | 分子名称: | 2-chloro-N-(2,2-dimethylpropyl)-N-[(3R)-1,1-dioxo-1lambda~6~-thiolan-3-yl]acetamide, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, SULFATE ION, ... | | 著者 | Seo, H.-S, Dhe-Paganon, S. | | 登録日 | 2019-12-17 | | 公開日 | 2020-12-30 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Sulfopin is a covalent inhibitor of Pin1 that blocks Myc-driven tumors in vivo.
Nat.Chem.Biol., 17, 2021
|
|
5AOC
 
 | | The structure of a novel thermophilic esterase from the Planctomycetes species, Thermogutta terrifontis, Est2-valerate bound | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Sayer, C, Szabo, Z, Isupov, M.N, Ingham, C, Littlechild, J.A. | | 登録日 | 2015-09-10 | | 公開日 | 2015-12-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | The Structure of a Novel Thermophilic Esterase from the Planctomycetes Species, Thermogutta Terrifontis Reveals an Open Active Site due to a Minimal 'CAP' Domain.
Front.Microbiol., 6, 2015
|
|
5AOA
 
 | | The structure of a novel thermophilic esterase from the Planctomycetes species, Thermogutta terrifontis, Est2-Propionate bound | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Sayer, C, Szabo, Z, Isupov, M.N, Ingham, C, Littlechild, J.A. | | 登録日 | 2015-09-10 | | 公開日 | 2015-12-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | The Structure of a Novel Thermophilic Esterase from the Planctomycetes Species, Thermogutta Terrifontis Reveals an Open Active Site due to a Minimal 'CAP' Domain.
Front.Microbiol., 6, 2015
|
|
5GS4
 
 | | Crystal structure of estrogen receptor alpha in complex with a stabilized peptide antagonist | | 分子名称: | ARG-IAS-ILE-LEU-DNP-ARG-LEU-LEU-GLN, ESTRADIOL, Estrogen receptor, ... | | 著者 | Xie, M, Wang, T, Li, Z.-G. | | 登録日 | 2016-08-13 | | 公開日 | 2017-08-30 | | 最終更新日 | 2018-07-18 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural Basis of Inhibition of ER alpha-Coactivator Interaction by High-Affinity N-Terminus Isoaspartic Acid Tethered Helical Peptides
J. Med. Chem., 60, 2017
|
|
5RPH
 
 | | PanDDA analysis group deposition -- Proteinase K changed state model for fragment Frag Xtal Screen A11a | | 分子名称: | 3-[3,4-bis(fluoranyl)phenyl]-1,4,6,7-tetrahydroimidazo[2,1-c][1,2,4]triazine, Proteinase K, SULFATE ION | | 著者 | Lima, G.M.A, Talibov, V, Benz, L.S, Jagudin, E, Mueller, U. | | 登録日 | 2020-09-23 | | 公開日 | 2021-05-26 | | 最終更新日 | 2021-06-23 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | FragMAXapp: crystallographic fragment-screening data-analysis and project-management system.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6V3C
 
 | |
5HDC
 
 | |
5HDD
 
 | |
5S20
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with PB1827975385 | | 分子名称: | (5R)-5-amino-5,6,7,8-tetrahydronaphthalen-1-ol, Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.037 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5HFO
 
 | | CRYSTAL STRUCTURE OF OXA-232 BETA-LACTAMASE | | 分子名称: | Beta-lactamase, CHLORIDE ION, GLYCEROL, ... | | 著者 | Retailleau, P, Oueslati, S, Cisse, C, Nordmann, P, Naas, T, Iorga, B. | | 登録日 | 2016-01-07 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.21 Å) | | 主引用文献 | Role of Arginine 214 in the Substrate Specificity of OXA-48.
Antimicrob.Agents Chemother., 64, 2020
|
|
6LXT
 
 | | Structure of post fusion core of 2019-nCoV S2 subunit | | 分子名称: | Spike protein S2, TETRAETHYLENE GLYCOL, ZINC ION | | 著者 | Zhu, Y, Sun, F. | | 登録日 | 2020-02-11 | | 公開日 | 2020-02-26 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion.
Cell Res., 30, 2020
|
|
5S4C
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z1954800348 | | 分子名称: | 1,4,5,6-tetrahydropyrimidin-2-amine, DIMETHYL SULFOXIDE, Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.01 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
6RLM
 
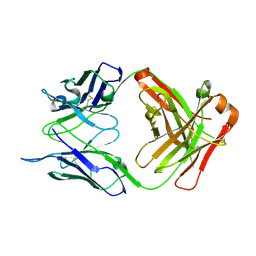 | | Crystal structure of AT1412dm Fab fragment | | 分子名称: | AT1412dm Fab (Heavy Chain), AT1412dm Fab (Light Chain), CHLORIDE ION | | 著者 | Neviani, V, Pearce, N.M, Pos, W, Schotte, R, Spits, H, Gros, P. | | 登録日 | 2019-05-02 | | 公開日 | 2021-05-12 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis of a homo-dimerization site in tetraspanin CD9 targeted by a melanoma patient-derived antibody
To Be Published
|
|
5S8Q
 
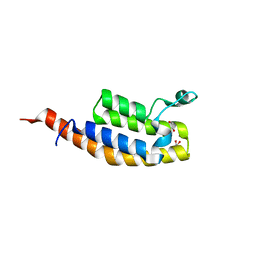 | | XChem group deposition -- Crystal Structure of the second bromodomain of pleckstrin homology domain interacting protein (PHIP) in complex with FMO3D000185a (space group P212121) | | 分子名称: | (3R)-3-methyl-1,2,3,4-tetrahydro-5H-1,4-benzodiazepin-5-one, 1,2-ETHANEDIOL, CALCIUM ION, ... | | 著者 | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F. | | 登録日 | 2020-12-17 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | XChem group deposition
To Be Published
|
|
5S2R
 
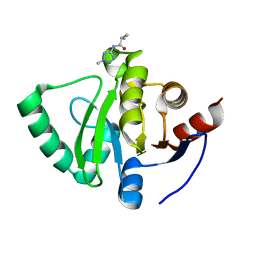 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with Z57292369 | | 分子名称: | 2-methyl-N-(2-methyl-2H-tetrazol-5-yl)propanamide, Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.132 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5DYW
 
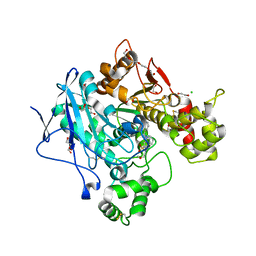 | | Crystal structure of human butyrylcholinesterase in complex with N-((1-benzylpiperidin-3-yl)methyl)-N-(2-methoxyethyl)naphthalene-2-sulfonamide | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Coquelle, N, Brus, B, Colletier, J.P. | | 登録日 | 2015-09-25 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Development of an in-vivo active reversible butyrylcholinesterase inhibitor.
Sci Rep, 6, 2016
|
|
5S3S
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with POB0103 | | 分子名称: | 1-[(5S,8R)-6,7,8,9-tetrahydro-5H-5,8-epiminocyclohepta[b]pyridin-10-yl]ethan-1-one, Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.039 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7MWS
 
 | | Crystal structure of tamarin CD81 large extracellular loop | | 分子名称: | CD81 protein, GLYCEROL, TETRAETHYLENE GLYCOL | | 著者 | Kumar, A, Hossain, R.A, Yost, S.A, Bu, W, Wang, Y, Dearborn, A.D, Grakoui, A, Cohen, J.I, Marcotrigiano, J. | | 登録日 | 2021-05-17 | | 公開日 | 2021-09-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural insights into hepatitis C virus receptor binding and entry.
Nature, 598, 2021
|
|
5HDS
 
 | |
5RWC
 
 | | INPP5D PanDDA analysis group deposition -- Crystal Structure of the phosphatase and C2 domains of SHIP1 in complex with Z2856434879 | | 分子名称: | 2-[(4-methyl-1H-imidazol-5-yl)methyl]-1,2,3,4-tetrahydroisoquinoline, DIMETHYL SULFOXIDE, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 | | 著者 | Bradshaw, W.J, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | 登録日 | 2020-10-30 | | 公開日 | 2020-11-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
7MZI
 
 | | SARS-CoV-2 receptor binding domain bound to Fab WCSL 129 | | 分子名称: | GLYCEROL, Spike protein S1, TETRAETHYLENE GLYCOL, ... | | 著者 | Pymm, P, Tan, L.L, Dietrich, M.H, Chan, L.J, Tham, W.H. | | 登録日 | 2021-05-24 | | 公開日 | 2021-10-06 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
6TPG
 
 | | Crystal structure of the Orexin-2 receptor in complex with EMPA at 2.74 A resolution | | 分子名称: | N-ethyl-2-[(6-methoxypyridin-3-yl)-(2-methylphenyl)sulfonyl-amino]-N-(pyridin-3-ylmethyl)ethanamide, OLEIC ACID, Orexin receptor type 2,GlgA glycogen synthase,Hypocretin receptor-2, ... | | 著者 | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | 登録日 | 2019-12-13 | | 公開日 | 2020-01-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.741 Å) | | 主引用文献 | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
6TQD
 
 | | D00-D0 domain of Intimin | | 分子名称: | BROMIDE ION, CHLORIDE ION, Intimin, ... | | 著者 | Weikum, J, Leo, J.C, Morth, J.P. | | 登録日 | 2019-12-16 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | The extracellular juncture domains in the intimin passenger adopt a constitutively extended conformation inducing restraints to its sphere of action.
Sci Rep, 10, 2020
|
|
