3DW4
 
 | | Crystal Structure of the Sarcin/Ricin Domain from E. COLI 23 S rRNA, U2650-OCH3 modified | | 分子名称: | GLYCEROL, Sarcin/Ricin Domain from E. Coli 23 S rRNA | | 著者 | Olieric, V, Rieder, U, Lang, K, Serganov, A, Schulze-Briese, C, Micura, R, Dumas, P, Ennifar, E. | | 登録日 | 2008-07-21 | | 公開日 | 2009-03-24 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (0.97 Å) | | 主引用文献 | A fast selenium derivatization strategy for crystallization and phasing of RNA structures.
Rna, 15, 2009
|
|
3PC6
 
 | |
3DZ4
 
 | | Human AdoMetDC with 5'-[(2-carboxamidoethyl)methylamino]-5'-deoxy-8-methyladenosine | | 分子名称: | 1,4-DIAMINOBUTANE, 3-[{[(2R,3S,4R,5R)-5-(6-amino-8-methyl-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}(methyl)amino]propanamid e, S-adenosylmethionine decarboxylase alpha chain, ... | | 著者 | Bale, S, McCloskey, D.E, Pegg, A.E, Secrist III, J.A, Guida, W.C, Ealick, S.E. | | 登録日 | 2008-07-29 | | 公開日 | 2009-03-10 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | New Insights into the Design of Inhibitors of Human S-Adenosylmethionine Decarboxylase: Studies of Adenine C8 Substitution in Structural Analogues of S-Adenosylmethionine
J.Med.Chem., 52, 2009
|
|
3PC7
 
 | |
4O0K
 
 | |
3PC4
 
 | |
3PDJ
 
 | | Crystal Structure of Human 11-beta-Hydroxysteroid Dehydrogenase 1 (11b-HSD1) in Complex with 4,4-Disubstituted Cyclohexylbenzamide Inhibitor | | 分子名称: | Corticosteroid 11-beta-dehydrogenase isozyme 1, N-[trans-4-(3-amino-3-oxopropyl)-4-phenylcyclohexyl]-N-cyclopropyl-4-[(1S)-2,2,2-trifluoro-1-hydroxy-1-methylethyl]benzamide, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Wang, Z, Sudom, A, Walker, N.P. | | 登録日 | 2010-10-22 | | 公開日 | 2011-10-05 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Synthesis and Optimization of Novel 4,4-Disubstituted Cyclohexylbenzamide Derivatives as Potent 11beta-HSD1 Inhibitors
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3DLQ
 
 | | Crystal structure of the IL-22/IL-22R1 complex | | 分子名称: | Interleukin-22, Interleukin-22 receptor subunit alpha-1 | | 著者 | Bleicher, L, de Moura, P.R, Watanabe, L, Colau, D, Dumoutier, L, Renauld, J.-C, Polikarpov, I. | | 登録日 | 2008-06-28 | | 公開日 | 2008-08-19 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of the IL-22/IL-22R1 complex and its implications for the IL-22 signaling mechanism
Febs Lett., 582, 2008
|
|
4O3Y
 
 | | Crystal structure of the vaccine antigen Transferrin Binding Protein B (TbpB) mutant Arg-179-Glu from Actinobacillus pleuropneumoniae H87 | | 分子名称: | ACETATE ION, GLYCEROL, Outer membrane protein; transferrin-binding protein | | 著者 | Calmettes, C, Yu, R.H, Schryvers, A.B, Moraes, T.F. | | 登録日 | 2013-12-18 | | 公開日 | 2015-01-14 | | 最終更新日 | 2015-03-04 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Nonbinding site-directed mutants of transferrin binding protein B exhibit enhanced immunogenicity and protective capabilities.
Infect.Immun., 83, 2015
|
|
3DMM
 
 | | Crystal structure of the CD8 alpha beta/H-2Dd complex | | 分子名称: | Beta-2 microglobulin, H-2 class I histocompatibility antigen, D-D alpha chain, ... | | 著者 | Wang, R, Natarajan, K, Margulies, D.H. | | 登録日 | 2008-07-01 | | 公開日 | 2009-07-14 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis of the CD8alphabeta/MHC class i interaction: focused recognition orients CD8beta to a T cell proximal position.
J.Immunol., 183, 2009
|
|
4O4X
 
 | | Crystal structure of the vaccine antigen Transferrin Binding Protein B (TbpB) double mutant Tyr-167-Ala and Trp-176-Ala from Haemophilus parasuis Hp5 | | 分子名称: | GLYCEROL, SODIUM ION, SULFATE ION, ... | | 著者 | Calmettes, C, Yu, R.H, Schryvers, A.B, Moraes, T.F. | | 登録日 | 2013-12-19 | | 公開日 | 2015-01-14 | | 最終更新日 | 2015-03-04 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Nonbinding site-directed mutants of transferrin binding protein B exhibit enhanced immunogenicity and protective capabilities.
Infect.Immun., 83, 2015
|
|
3PDC
 
 | |
4NLO
 
 | |
3PC2
 
 | |
4NN1
 
 | |
4JR4
 
 | | Crystal structure of Mtb DsbA (Oxidized) | | 分子名称: | Possible conserved membrane or secreted protein, SULFATE ION | | 著者 | Wang, L. | | 登録日 | 2013-03-21 | | 公開日 | 2013-07-17 | | 最終更新日 | 2017-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.498 Å) | | 主引用文献 | Structure analysis of the extracellular domain reveals disulfide bond forming-protein properties of Mycobacterium tuberculosis Rv2969c.
Protein Cell, 4, 2013
|
|
5RSJ
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000089254160_N3 | | 分子名称: | 3-[(2-methyl-1,3-thiazol-4-yl)methyl]-3H-purin-6-amine, Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
4O5R
 
 | | Crystal structure of Alkylhydroperoxide Reductase subunit C from E. coli | | 分子名称: | AhpC component, subunit of alkylhydroperoxide reductase | | 著者 | Dip, P.V, Kamariah, N, Manimekalai, M.S.S, Gruber, G, Eisenhaber, F, Eisenhaber, B. | | 登録日 | 2013-12-20 | | 公開日 | 2014-11-05 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.33 Å) | | 主引用文献 | Structure, mechanism and ensemble formation of the alkylhydroperoxide reductase subunits AhpC and AhpF from Escherichia coli
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5RT0
 
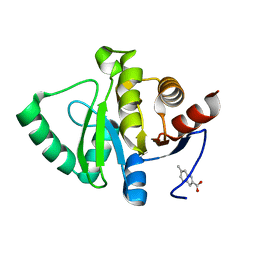 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000002582714 | | 分子名称: | 6-methyl-1H-indole-2-carboxylic acid, Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RTH
 
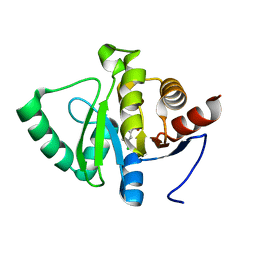 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000156863 | | 分子名称: | 3-chlorobenzoate, Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
4JSP
 
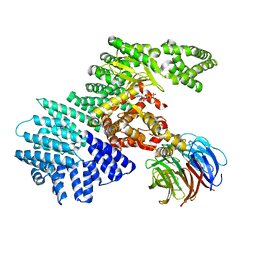 | |
3DN7
 
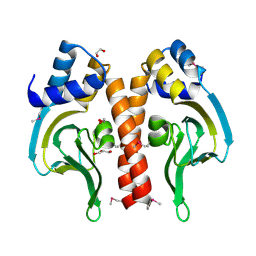 | | Cyclic nucleotide binding regulatory protein from Cytophaga hutchinsonii. | | 分子名称: | 1,2-ETHANEDIOL, Cyclic nucleotide binding regulatory protein, GLYCEROL | | 著者 | Osipiuk, J, Maltseva, N, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2008-07-01 | | 公開日 | 2008-08-26 | | 最終更新日 | 2017-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | X-ray crystal structure of cyclic nucleotide binding regulatory protein from Cytophaga hutchinsonii.
To be Published
|
|
3O86
 
 | | Crystal structure of AmpC beta-lactamase in complex with a sulfonamide boronic acid inhibitor | | 分子名称: | Beta-lactamase, PHOSPHATE ION, {[(benzylsulfonyl)amino]methyl}boronic acid | | 著者 | Eidam, O, Romagnoli, C, Karpiak, J, Shoichet, B.K. | | 登録日 | 2010-08-02 | | 公開日 | 2010-11-03 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Design, Synthesis, Crystal Structures, and Antimicrobial Activity of Sulfonamide Boronic Acids as beta-Lactamase Inhibitors
J.Med.Chem., 53, 2010
|
|
5RTZ
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000404062 | | 分子名称: | 3-FLUORO-4-HYDROXYBENZOIC ACID, Non-structural protein 3 | | 著者 | Correy, G.J, Young, I.D, Thompson, M.C, Fraser, J.S. | | 登録日 | 2020-09-28 | | 公開日 | 2020-12-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
3O8R
 
 | |
