7ZA3
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | 著者 | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | 登録日 | 2022-03-21 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (4 Å) | | 主引用文献 | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZAW
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3 | | 著者 | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | 登録日 | 2022-03-22 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
7ZA2
 
 | | GPC3-Unc5D octamer structure and role in cell migration | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glypican-3, ... | | 著者 | Akkermans, O, Delloye-Bourgeois, C, Peregrina, C, Carrasquero, M, Kokolaki, M, Berbeira-Santana, M, Chavent, M, Reynaud, F, Ritu, R, Agirre, J, Aksu, M, White, E, Lowe, E, Ben Amar, D, Zaballa, S, Huo, J, Pakos, I, McCubbin, P, Comoletti, D, Owens, R, Robinson, C, Castellani, V, del Toro, D, Seiradake, E. | | 登録日 | 2022-03-21 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (4.6 Å) | | 主引用文献 | GPC3-Unc5 receptor complex structure and role in cell migration.
Cell, 185, 2022
|
|
4TTM
 
 | |
5HG9
 
 | | EGFR (L858R, T790M, V948R) in complex with 1-[(3R,4R)-3-[({2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-(trifluoromethyl)pyrrolidin-1-yl]prop-2-en-1-one | | 分子名称: | 1-[(3R,4R)-3-[({2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-(trifluoromethyl)pyrrolidin-1-yl]propan-1-one, Epidermal growth factor receptor, GLYCEROL, ... | | 著者 | Gajiwala, K.S. | | 登録日 | 2016-01-08 | | 公開日 | 2016-02-03 | | 最終更新日 | 2016-03-23 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Discovery of 1-{(3R,4R)-3-[({5-Chloro-2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-methoxypyrrolidin-1-yl}prop-2-en-1-one (PF-06459988), a Potent, WT Sparing, Irreversible Inhibitor of T790M-Containing EGFR Mutants.
J.Med.Chem., 59, 2016
|
|
6RNL
 
 | | L-[Ru(TAP)2(dppz)]2+ bound to the G-quadruplex forming sequence d(TAGGGTT) | | 分子名称: | DNA (5'-D(*TP*AP*GP*GP*GP*TP*T)-3'), POTASSIUM ION, Ru(tap)2(dppz) complex, ... | | 著者 | McQuaid, K.T, Hall, J.P, Cardin, C.J. | | 登録日 | 2019-05-09 | | 公開日 | 2019-08-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Three thymine/adenine binding modes of the ruthenium complex Lambda-[Ru(TAP)2(dppz)]2+to the G-quadruplex forming sequence d(TAGGGTT) shown by X-ray crystallography.
Chem.Commun.(Camb.), 55, 2019
|
|
7LST
 
 | | Ruminococcus bromii Amy12-D392A with 63-a-D-glucosyl-maltotriosyl-maltotriose | | 分子名称: | ACETATE ION, CALCIUM ION, GLYCEROL, ... | | 著者 | Koropatkin, N.M, Cockburn, D.W, Brown, H.A, Kibler, R.D. | | 登録日 | 2021-02-18 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structure and substrate recognition by the Ruminococcus bromii amylosome pullulanases.
J.Struct.Biol., 213, 2021
|
|
7LSU
 
 | | Ruminococcus bromii Amy12-D392A with 63-a-D-glucosyl-maltotriose | | 分子名称: | ACETATE ION, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Koropatkin, N.M, Cockburn, D.W, Brown, H.A, Kibler, R.D. | | 登録日 | 2021-02-18 | | 公開日 | 2021-07-14 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure and substrate recognition by the Ruminococcus bromii amylosome pullulanases.
J.Struct.Biol., 213, 2021
|
|
6V82
 
 | | Crystal structure of tryptophan synthase from Chlamydia trachomatis D/UW-3/CX | | 分子名称: | SULFATE ION, Tryptophan synthase alpha chain, Tryptophan synthase beta chain, ... | | 著者 | Michalska, K, Maltseva, N, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-12-10 | | 公開日 | 2020-12-16 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.424 Å) | | 主引用文献 | Catalytically impaired TrpA subunit of tryptophan synthase from Chlamydia trachomatis is an allosteric regulator of TrpB.
Protein Sci., 30, 2021
|
|
4XSM
 
 | | Crystal structure of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with D-talitol | | 分子名称: | D-altritol, D-tagatose 3-epimerase, MANGANESE (II) ION | | 著者 | Yoshida, H, Yoshihara, A, Ishii, T, Izumori, K, Kamitori, S. | | 登録日 | 2015-01-22 | | 公開日 | 2016-01-27 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
5HG7
 
 | | EGFR (L858R, T790M, V948R) in complex with 1-{(3R,4R)-3-[5-Chloro-2-(1-methyl-1H-pyrazol-4-ylamino)-7H-pyrrolo[2,3-d]pyrimidin-4-yloxymethyl]-4-methoxy-pyrrolidin-1-yl}propenone (PF-06459988) | | 分子名称: | 1-{(3R,4R)-3-[({5-chloro-2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-methoxypyrrolidin-1-yl}propan-1-one, Epidermal growth factor receptor, SULFATE ION | | 著者 | Gajiwala, K.S. | | 登録日 | 2016-01-08 | | 公開日 | 2016-01-27 | | 最終更新日 | 2016-03-23 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Discovery of 1-{(3R,4R)-3-[({5-Chloro-2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-methoxypyrrolidin-1-yl}prop-2-en-1-one (PF-06459988), a Potent, WT Sparing, Irreversible Inhibitor of T790M-Containing EGFR Mutants.
J.Med.Chem., 59, 2016
|
|
5HG8
 
 | | EGFR (L858R, T790M, V948R) in complex with N-[3-({2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)phenyl]prop-2-enamide | | 分子名称: | Epidermal growth factor receptor, GLYCEROL, N-[3-({2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)phenyl]propanamide, ... | | 著者 | Gajiwala, K.S. | | 登録日 | 2016-01-08 | | 公開日 | 2016-02-03 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Discovery of 1-{(3R,4R)-3-[({5-Chloro-2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-methoxypyrrolidin-1-yl}prop-2-en-1-one (PF-06459988), a Potent, WT Sparing, Irreversible Inhibitor of T790M-Containing EGFR Mutants.
J.Med.Chem., 59, 2016
|
|
6T2H
 
 | | Furano[2,3-d]prymidine amides as Notum inhibitors | | 分子名称: | 1,2-ETHANEDIOL, 2-[[(4~{S})-5-chloranyl-6-methyl-1,2,3,4-tetrahydrothieno[2,3-d]pyrimidin-4-yl]sulfanyl]ethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhao, Y, Jones, E.Y. | | 登録日 | 2019-10-08 | | 公開日 | 2020-01-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | Scaffold-hopping identifies furano[2,3-d]pyrimidine amides as potent Notum inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
5HG5
 
 | | EGFR (L858R, T790M, V948R) in complex with N-{3-[(2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-7H-pyrrolo[2,3-d]pyrimidin-4-yl)oxy]phenyl}prop-2-enamide | | 分子名称: | Epidermal growth factor receptor, GLYCEROL, N-{3-[(2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-7H-pyrrolo[2,3-d]pyrimidin-4-yl)oxy]phenyl}propanamide, ... | | 著者 | Gajiwala, K.S. | | 登録日 | 2016-01-08 | | 公開日 | 2016-02-03 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Discovery of 1-{(3R,4R)-3-[({5-Chloro-2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-methoxypyrrolidin-1-yl}prop-2-en-1-one (PF-06459988), a Potent, WT Sparing, Irreversible Inhibitor of T790M-Containing EGFR Mutants.
J.Med.Chem., 59, 2016
|
|
4PPR
 
 | | Crystal structure of Mycobacterium tuberculosis D,D-peptidase Rv3330 in complex with meropenem | | 分子名称: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Penicillin-binding protein DacB1 | | 著者 | Prigozhin, D.M, Huizar, J.P, Mavrici, D, Alber, T, TB Structural Genomics Consortium (TBSGC) | | 登録日 | 2014-02-27 | | 公開日 | 2014-11-05 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Subfamily-specific adaptations in the structures of two penicillin-binding proteins from Mycobacterium tuberculosis.
Plos One, 9, 2014
|
|
6PF6
 
 | | Crystal structure of TS-DHFR from Cryptosporidium hominis in complex with NADPH, FdUMP and 2-(4-((2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl)benzamido)terephthalic acid | | 分子名称: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-({4-[(2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl]benzene-1-carbonyl}amino)benzene-1,4-dicarboxylic acid, Thymidylate synthase,Thymidylate synthase | | 著者 | Czyzyk, D.L, Valhondo, M, Jorgensen, W.L, Anderson, K.S. | | 登録日 | 2019-06-21 | | 公開日 | 2019-10-16 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.502 Å) | | 主引用文献 | Structure activity relationship towards design of cryptosporidium specific thymidylate synthase inhibitors.
Eur.J.Med.Chem., 183, 2019
|
|
4Y0T
 
 | | Crystal structure of apo form of OXA-58, a Carbapenem hydrolyzing Class D beta-lactamase from Acinetobacter baumanii (P21, 4mol/ASU) | | 分子名称: | Beta-lactamase | | 著者 | Pratap, S, Katiki, M, Gill, P, Golemi-Kotra, D, Kumar, P. | | 登録日 | 2015-02-06 | | 公開日 | 2016-01-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Active-Site Plasticity Is Essential to Carbapenem Hydrolysis by OXA-58 Class D beta-Lactamase of Acinetobacter baumannii.
Antimicrob.Agents Chemother., 60, 2015
|
|
6P9D
 
 | | Crystal Structure of Pseudomonas aeruginosa D-Arginine Dehydrogenase Y249F variant with FAD - Yellow fraction | | 分子名称: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, FAD-dependent catabolic D-arginine dehydrogenase DauA, GLYCEROL | | 著者 | Reis, R.A.G, Iyer, A, Agniswamy, J, Gannavaram, S, Weber, I, Gadda, G. | | 登録日 | 2019-06-10 | | 公開日 | 2020-06-17 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.329 Å) | | 主引用文献 | A Single-Point Mutation in d-Arginine Dehydrogenase Unlocks a Transient Conformational State Resulting in Altered Cofactor Reactivity.
Biochemistry, 60, 2021
|
|
6PF5
 
 | | Crystal structure of human thymidylate synthase Delta (7-29) in complex with dUMP and 2-(2-(4-((2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl)benzamido)-4-methoxyphenyl)acetic acid | | 分子名称: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase, [2-({4-[(2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl]benzene-1-carbonyl}amino)-4-methoxyphenyl]acetic acid | | 著者 | Czyzyk, D.J, Valhondo, M, Jorgensen, W.L, Anderson, K.S. | | 登録日 | 2019-06-21 | | 公開日 | 2019-10-02 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Structure activity relationship towards design of cryptosporidium specific thymidylate synthase inhibitors.
Eur.J.Med.Chem., 183, 2019
|
|
5FHJ
 
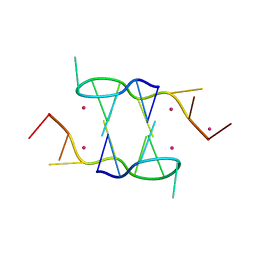 | |
4Y0U
 
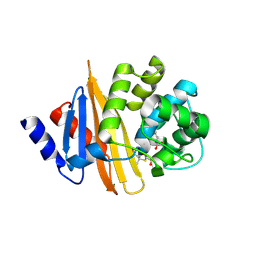 | | Crystal Structure of 6Alpha-Hydroxymethylpenicillanate Complexed with OXA-58, a Carbapenem hydrolyzing Class D betalactamase from Acinetobacter baumanii. | | 分子名称: | 2-(1-CARBOXY-2-HYDROXY-ETHYL)-5,5-DIMETHYL-THIAZOLIDINE-4-CARBOXYLIC ACID, Beta-lactamase | | 著者 | Pratap, S, Katiki, M, Gill, P, Golemi-Kotra, D, Kumar, P. | | 登録日 | 2015-02-06 | | 公開日 | 2016-01-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Active-Site Plasticity Is Essential to Carbapenem Hydrolysis by OXA-58 Class D beta-Lactamase of Acinetobacter baumannii.
Antimicrob.Agents Chemother., 60, 2015
|
|
8E75
 
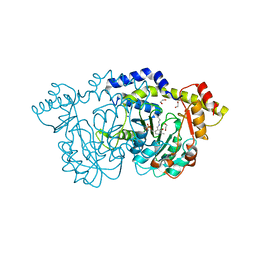 | | Crystal structure of Pcryo_0616, the aminotransferase required to synthesize UDP-N-acetyl-3-amino-D-glucosaminuronic acid (UDP-GlcNAc3NA) | | 分子名称: | 1,2-ETHANEDIOL, DegT/DnrJ/EryC1/StrS aminotransferase, SODIUM ION | | 著者 | Hofmeister, D.L, Seltzner, C.A, Bockhaus, N.J, thoden, J.B, Holden, H.M. | | 登録日 | 2022-08-23 | | 公開日 | 2022-11-23 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Investigation of the enzymes required for the biosynthesis of 2,3-diacetamido-2,3-dideoxy-d-glucuronic acid in Psychrobacter cryohalolentis K5 T.
Protein Sci., 32, 2023
|
|
4Q3I
 
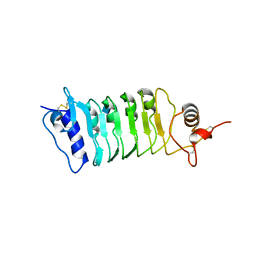 | | Structure of the OsSERK2 leucine rich repeat extracellular domain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, OsSERK2 D128N | | 著者 | McAndrew, R.P, Pruitt, R.N, Kamita, S.G, Pereira, J.H, Majumder, D, Hammock, B.D, Adams, P.D, Ronald, P.C. | | 登録日 | 2014-04-11 | | 公開日 | 2014-11-12 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structure of the OsSERK2 leucine-rich repeat extracellular domain.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4Q3G
 
 | | Structure of the OsSERK2 leucine rich repeat extracellular domain | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, OsSERK2 | | 著者 | McAndrew, R.P, Pruitt, R.N, Kamita, S.G, Pereira, J.H, Majumder, D, Hammock, B.D, Adams, P.D, Ronald, P.C. | | 登録日 | 2014-04-11 | | 公開日 | 2014-11-12 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.787 Å) | | 主引用文献 | Structure of the OsSERK2 leucine-rich repeat extracellular domain.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4Z06
 
 | | C. bescii Family 3 pectate lyase double mutant K108A/R133A in complex with ALPHA-D-GALACTOPYRANURONIC ACID | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | 著者 | Alahuhta, P.M, Lunin, V.V. | | 登録日 | 2015-03-25 | | 公開日 | 2015-12-23 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | The catalytic mechanism and unique low pH optimum of Caldicellulosiruptor bescii family 3 pectate lyase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
