8YRT
 
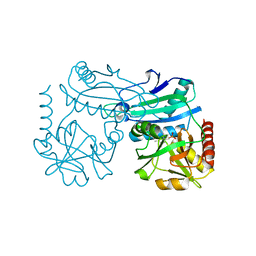 | | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis in the holo form obtained at pH 7.0 | | 分子名称: | Aminotransferase class IV, PYRIDOXAL-5'-PHOSPHATE | | 著者 | Matyuta, I.O, Bakunova, A.K, Minyaev, M.E, Popov, V.O, Boyko, K.M. | | 登録日 | 2024-03-21 | | 公開日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of D-amino acid transaminase from Haliscomenobacter hydrossis in the holo form obtained at pH 7.0
To Be Published
|
|
8DL5
 
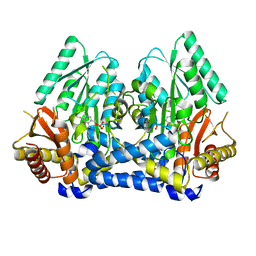 | |
7VMY
 
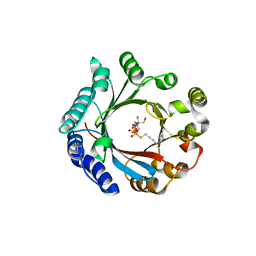 | | Crystal structure of LimF prenyltransferase bound with GSPP | | 分子名称: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GERANYL S-THIOLODIPHOSPHATE, LynF/TruF/PatF family peptide O-prenyltransferase, ... | | 著者 | Hamada, K, Kobayashi, S, Okada, C, Zhang, Y, Inoue, S, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | 登録日 | 2021-10-09 | | 公開日 | 2022-08-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | LimF is a versatile prenyltransferase for histidine-C-geranylation on diverse non-natural substrates
Nat Catal, 2022
|
|
7VMW
 
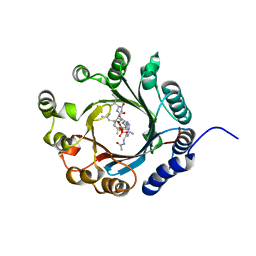 | | Crystal structure of LimF prenyltransferase bound with a peptide substrate and GSPP | | 分子名称: | GERANYL S-THIOLODIPHOSPHATE, LynF/TruF/PatF family peptide O-prenyltransferase, MAGNESIUM ION, ... | | 著者 | Hamada, K, Kobayashi, S, Okada, C, Zhang, Y, Inoue, S, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | 登録日 | 2021-10-09 | | 公開日 | 2022-08-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | LimF is a versatile prenyltransferase for histidine-C-geranylation on diverse non-natural substrates
Nat Catal, 2022
|
|
4GPA
 
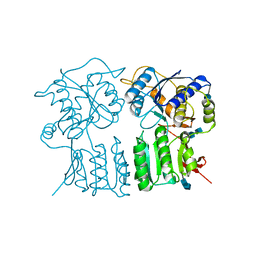 | |
8JE4
 
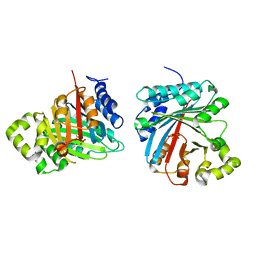 | | Crystal structure of LimF prenyltransferase (H239G/W273T mutant) bound with the thiodiphosphate moiety of farnesyl S-thiolodiphosphate (FSPP) | | 分子名称: | MAGNESIUM ION, TRIHYDROGEN THIODIPHOSPHATE, prenyltransferase, ... | | 著者 | Hamada, K, Oguni, A, Zhang, Y, Satake, M, Goto, Y, Suga, H, Ogata, K, Sengoku, T. | | 登録日 | 2023-05-15 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | Switching Prenyl Donor Specificities of Cyanobactin Prenyltransferases.
J.Am.Chem.Soc., 145, 2023
|
|
8KDL
 
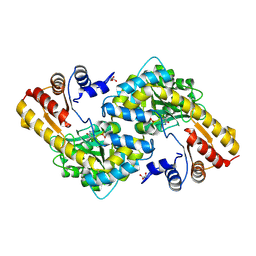 | |
8Q3V
 
 | | Cryo-EM structure of the methanogenic Na+ translocating N5-methyl-H4MPT:CoM methyltransferase complex | | 分子名称: | MAGNESIUM ION, SODIUM ION, Tetrahydromethanopterin S-methyltransferase subunit A 1, ... | | 著者 | Aziz, I, Vonck, J, Ermler, U. | | 登録日 | 2023-08-04 | | 公開日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (2.08 Å) | | 主引用文献 | Structural and mechanistic basis of the central energy-converting methyltransferase complex of methanogenesis.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8Q54
 
 | |
2CMO
 
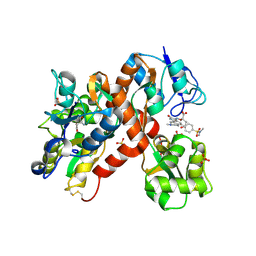 | | The structure of a mixed glur2 ligand-binding core dimer in complex with (s)-glutamate and the antagonist (s)-ns1209 | | 分子名称: | 2-({[(3E)-5-{4-[(DIMETHYLAMINO)(DIHYDROXY)-LAMBDA~4~-SULFANYL]PHENYL}-8-METHYL-2-OXO-6,7,8,9-TETRAHYDRO-1H-PYRROLO[3,2-H]ISOQUINOLIN-3(2H)-YLIDENE]AMINO}OXY)-4-HYDROXYBUTANOIC ACID, GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, ... | | 著者 | Kasper, C, Pickering, D.S, Mirza, O, Olsen, L, Kristensen, A.S, Greenwood, J.R, Liljefors, T, Schousboe, A, Watjen, F, Gajhede, M, Sigurskjold, B.W, Kastrup, J.S. | | 登録日 | 2006-05-11 | | 公開日 | 2006-06-06 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | The Structure of a Mixed Glur2 Ligand-Binding Core Dimer in Complex with (S)-Glutamate and the Antagonist (S)-Ns1209.
J.Mol.Biol., 357, 2006
|
|
1N0T
 
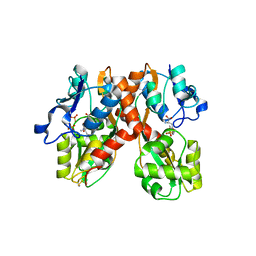 | | X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with the antagonist (S)-ATPO at 2.1 A resolution. | | 分子名称: | (S)-2-AMINO-3-(5-TERT-BUTYL-3-(PHOSPHONOMETHOXY)-4-ISOXAZOLYL)PROPIONIC ACID, ACETATE ION, Glutamate receptor 2, ... | | 著者 | Hogner, A, Greenwood, J.R, Liljefors, T, Lunn, M.-L, Egebjerg, J, Larsen, I.K, Gouaux, E, Kastrup, J.S. | | 登録日 | 2002-10-15 | | 公開日 | 2003-03-04 | | 最終更新日 | 2017-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Competitive antagonism of AMPA receptors by ligands of
different classes: crystal structure of ATPO bound to the
GluR2 ligand-binding core, in comparison with DNQX.
J.Med.Chem., 46, 2003
|
|
1XHY
 
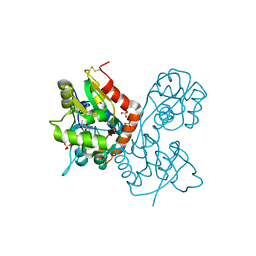 | | X-ray structure of the Y702F mutant of the GluR2 ligand-binding core (S1S2J) in complex with kainate at 1.85 A resolution | | 分子名称: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor, SULFATE ION | | 著者 | Frandsen, A, Pickering, D.S, Vestergaard, B, Kasper, C, Nielsen, B.B, Greenwood, J.R, Campiani, G, Gajhede, M, Schousboe, A, Kastrup, J.S. | | 登録日 | 2004-09-21 | | 公開日 | 2005-03-22 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Tyr702 Is an Important Determinant of Agonist Binding and Domain Closure of the Ligand-Binding Core of GluR2.
Mol.Pharmacol., 67, 2005
|
|
8TKP
 
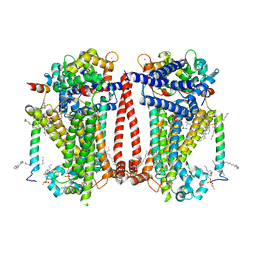 | | Structure of the C. elegans TMC-2 complex | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Clark, S, Jeong, H, Goehring, A, Posert, R, Gouaux, E. | | 登録日 | 2023-07-25 | | 公開日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | The structure of the Caenorhabditis elegans TMC-2 complex suggests roles of lipid-mediated subunit contacts in mechanosensory transduction.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
1JBM
 
 | |
1HBM
 
 | | METHYL-COENZYME M REDUCTASE ENZYME PRODUCT COMPLEX | | 分子名称: | CHLORIDE ION, FACTOR 430, GLYCEROL, ... | | 著者 | Ermler, U, Grabarse, W. | | 登録日 | 2001-04-20 | | 公開日 | 2001-08-16 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | On the Mechanism of Biological Methane Formation: Structural Evidence for Conformational Changes in Methyl-Coenzyme M Reductase Upon Substrate Binding
J.Mol.Biol., 309, 2001
|
|
1HBU
 
 | |
1HBO
 
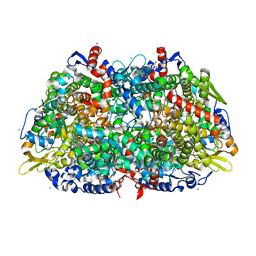 | | METHYL-COENZYME M REDUCTASE MCR-RED1-SILENT | | 分子名称: | 1-THIOETHANESULFONIC ACID, CHLORIDE ION, Coenzyme B, ... | | 著者 | Grabarse, W. | | 登録日 | 2001-04-20 | | 公開日 | 2001-08-16 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | On the Mechanism of Biological Methane Formation: Structural Evidence for Conformational Changes in Methyl-Coenzyme M Reductase Upon Substrate Binding
J.Mol.Biol., 309, 2001
|
|
1HBN
 
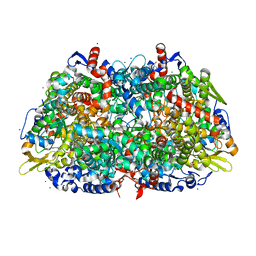 | | METHYL-COENZYME M REDUCTASE | | 分子名称: | 1-THIOETHANESULFONIC ACID, CHLORIDE ION, Coenzyme B, ... | | 著者 | Ermler, U, Grabarse, W. | | 登録日 | 2001-04-20 | | 公開日 | 2001-08-16 | | 最終更新日 | 2019-07-24 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | On the Mechanism of Biological Methane Formation: Structural Evidence for Conformational Changes in Methyl-Coenzyme M Reductase Upon Substrate Binding
J.Mol.Biol., 309, 2001
|
|
1SYH
 
 | | X-RAY STRUCTURE OF THE GLUR2 LIGAND-BINDING CORE (S1S2J) IN COMPLEX WITH (S)-CPW399 AT 1.85 A RESOLUTION. | | 分子名称: | (S)-2-AMINO-3-(1,3,5,7-PENTAHYDRO-2,4-DIOXO-CYCLOPENTA[E]PYRIMIDIN-1-YL) PROIONIC ACID, Glutamate receptor 2 | | 著者 | Frandsen, A, Pickering, D.S, Vestergaard, B, Kasper, C, Nielsen, B.B, Greenwood, J.R, Campiani, G, Gajhede, M, Schousboe, A, Kastrup, J.S. | | 登録日 | 2004-04-01 | | 公開日 | 2005-03-22 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Tyr702 Is an Important Determinant of Agonist Binding and Domain Closure of the Ligand-Binding Core of GluR2.
Mol.Pharmacol., 67, 2005
|
|
1SYI
 
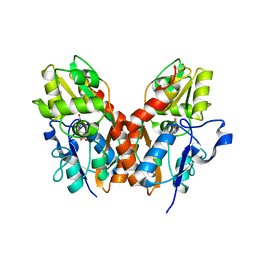 | | X-RAY STRUCTURE OF THE Y702F MUTANT OF THE GLUR2 LIGAND-BINDING CORE (S1S2J) IN COMPLEX WITH (S)-CPW399 AT 2.1 A RESOLUTION. | | 分子名称: | (S)-2-AMINO-3-(1,3,5,7-PENTAHYDRO-2,4-DIOXO-CYCLOPENTA[E]PYRIMIDIN-1-YL) PROIONIC ACID, Glutamate receptor 2 | | 著者 | Frandsen, A, Pickering, D.S, Vestergaard, B, Kasper, C, Nielsen, B.B, Greenwood, J.R, Campiani, G, Gajhede, M, Schousboe, A, Kastrup, J.S. | | 登録日 | 2004-04-01 | | 公開日 | 2005-03-22 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Tyr702 Is an Important Determinant of Agonist Binding and Domain Closure of the Ligand-Binding Core of GluR2.
Mol.Pharmacol., 67, 2005
|
|
2KKE
 
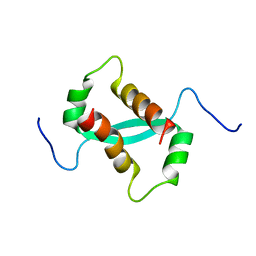 | | Solution NMR Structure of a dimeric protein of unknown function from Methanobacterium thermoautotrophicum, Northeast Structural Genomics Consortium Target TR5 | | 分子名称: | Uncharacterized protein | | 著者 | Swapna, G.V.T, Gunsalus, X, Huang, L, Xiao, K, Everett, J.K, Acton, T.B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2009-06-18 | | 公開日 | 2009-07-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Solution Structure of a putative uncharacterized protein from Methanobacterium thermoautotrophicum, Northeast Structural Genomics Consortium Target:TR5
To be Published
|
|
1NHO
 
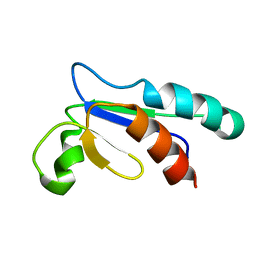 | | Structural and Functional characterization of a Thioredoxin-like Protein from Methanobacterium thermoautotrophicum | | 分子名称: | Probable Thioredoxin | | 著者 | Amegbey, G.Y, Monzavi, H, Habibi-Nazhad, B, Bhattacharyya, S, Wishart, D.S. | | 登録日 | 2002-12-19 | | 公開日 | 2003-08-26 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural and Functional Characterization of a Thioredoxin-like Protein (Mt0807) from Methanobacterium thermoautotrophicum
Biochemistry, 42, 2003
|
|
5IPU
 
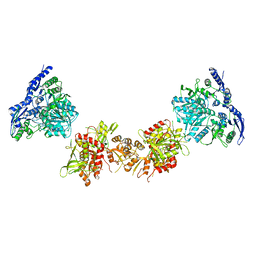 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the DCKA/D-APV-bound conformation, state 6 | | 分子名称: | Ionotropic glutamate receptor subunit NR2B, N-methyl-D-aspartate receptor subunit NR1-8a | | 著者 | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, Mchaourab, S.H, Gouaux, E. | | 登録日 | 2016-03-09 | | 公開日 | 2016-04-20 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (15.4 Å) | | 主引用文献 | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
5IPT
 
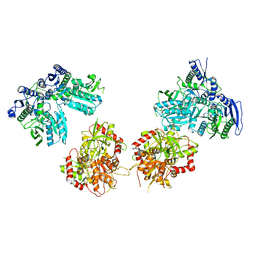 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the DCKA/D-APV-bound conformation, state 5 | | 分子名称: | Ionotropic glutamate receptor subunit NR2B, N-methyl-D-aspartate receptor subunit NR1-8a | | 著者 | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, Mchaourab, S.H, Gouaux, E. | | 登録日 | 2016-03-09 | | 公開日 | 2016-04-20 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (14.1 Å) | | 主引用文献 | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
5IPS
 
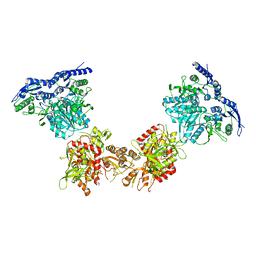 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the DCKA/D-APV-bound conformation, state 4 | | 分子名称: | Ionotropic glutamate receptor subunit NR2B, N-methyl-D-aspartate receptor subunit NR1-8a | | 著者 | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, Mchaourab, S.H, Gouaux, E. | | 登録日 | 2016-03-09 | | 公開日 | 2016-04-20 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (13.5 Å) | | 主引用文献 | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
