8FWT
 
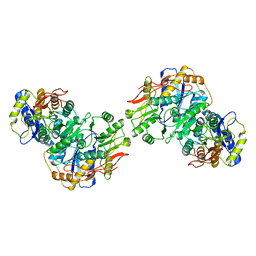 | | Structure of the amino terminal domain of kainate receptor GluK2 in complex with the positive allosteric modulator BPAM344 and competitive antagonist DNQX | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | 著者 | Yen, L.Y, Gangwar, S.P, Yelshanskaya, M.V, Sobolevsky, A.I. | | 登録日 | 2023-01-23 | | 公開日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (3.09 Å) | | 主引用文献 | Positive and negative allosteric modulation of GluK2 kainate receptors by BPAM344 and antiepileptic perampanel.
Cell Rep, 42, 2023
|
|
8FWQ
 
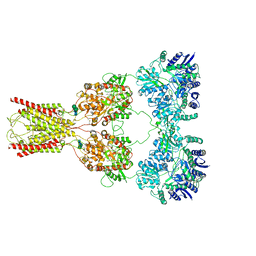 | | Structure of kainate receptor GluK2 in complex with the positive allosteric modulator BPAM344 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-cyclopropyl-7-fluoro-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, ... | | 著者 | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | 登録日 | 2023-01-23 | | 公開日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (3.96 Å) | | 主引用文献 | Positive and negative allosteric modulation of GluK2 kainate receptors by BPAM344 and antiepileptic perampanel.
Cell Rep, 42, 2023
|
|
8FWW
 
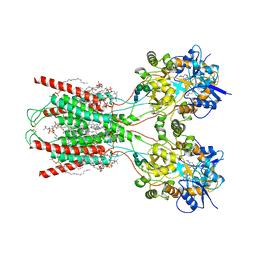 | | Structure of the ligand-binding and transmembrane domains of kainate receptor GluK2 in complex with the positive allosteric modulator BPAM344 and noncompetitive inhibitor perampanel | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-(6'-oxo-1'-phenyl[1',6'-dihydro[2,3'-bipyridine]]-5'-yl)benzonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | 登録日 | 2023-01-23 | | 公開日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Positive and negative allosteric modulation of GluK2 kainate receptors by BPAM344 and antiepileptic perampanel.
Cell Rep, 42, 2023
|
|
8FWR
 
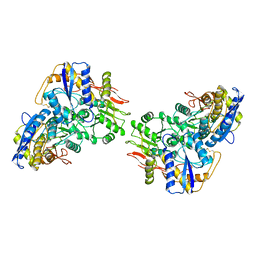 | | Structure of the amino-terminal domain of kainate receptor GluK2 in complex with the positive allosteric modulator BPAM344 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, kainate 2, ... | | 著者 | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | 登録日 | 2023-01-23 | | 公開日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Positive and negative allosteric modulation of GluK2 kainate receptors by BPAM344 and antiepileptic perampanel.
Cell Rep, 42, 2023
|
|
8FWV
 
 | | Structure of the amino-terminal domain of kainate receptor GluK2 in complex with the positive allosteric modulator BPAM344 and noncompetitive inhibitor perampanel | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | 著者 | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | 登録日 | 2023-01-23 | | 公開日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (3.03 Å) | | 主引用文献 | Positive and negative allosteric modulation of GluK2 kainate receptors by BPAM344 and antiepileptic perampanel.
Cell Rep, 42, 2023
|
|
8FWU
 
 | | Structure of the ligand-binding and transmembrane domains of kainate receptor GluK2 in complex with the positive allosteric modulator BPAM344 and competitive antagonist DNQX | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Yen, L.Y, Gangwar, S.P, Yelshanskaya, M.V, Sobolevsky, A.I. | | 登録日 | 2023-01-23 | | 公開日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Positive and negative allosteric modulation of GluK2 kainate receptors by BPAM344 and antiepileptic perampanel.
Cell Rep, 42, 2023
|
|
7FDJ
 
 | | Engineered Hepatitis B virus core antigen with short linker T=4 | | 分子名称: | Capsid protein,Immunoglobulin G-binding protein A | | 著者 | Jeong, H, Heo, Y, Yoo, Y, Ryu, B, Yun, J, Cho, H, Lee, W. | | 登録日 | 2021-07-16 | | 公開日 | 2021-09-01 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Structural and Functional Characterizations of Cancer Targeting Nanoparticles Based on Hepatitis B Virus Capsid.
Int J Mol Sci, 22, 2021
|
|
7EP6
 
 | | Engineered Hepatitis B virus core antigen T=4 | | 分子名称: | Capsid protein,Immunoglobulin G-binding protein A | | 著者 | Jeong, H, Heo, Y, Yoo, Y, Ryu, B, Yun, J, Cho, H, Lee, W. | | 登録日 | 2021-04-26 | | 公開日 | 2021-09-01 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (3.86 Å) | | 主引用文献 | Structural and Functional Characterizations of Cancer Targeting Nanoparticles Based on Hepatitis B Virus Capsid.
Int J Mol Sci, 22, 2021
|
|
7MXO
 
 | | CryoEM structure of human NKCC1 | | 分子名称: | CHLORIDE ION, POTASSIUM ION, Solute carrier family 12 member 2 | | 著者 | Moseng, M.A. | | 登録日 | 2021-05-19 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-05-31 | | 実験手法 | ELECTRON MICROSCOPY (3.47 Å) | | 主引用文献 | Inhibition mechanism of NKCC1 involves the carboxyl terminus and long-range conformational coupling.
Sci Adv, 8, 2022
|
|
7N3N
 
 | | CryoEM structure of human NKCC1 state Fu-I | | 分子名称: | 5-(AMINOSULFONYL)-4-CHLORO-2-[(2-FURYLMETHYL)AMINO]BENZOIC ACID, Solute carrier family 12 member 2 | | 著者 | Moseng, M.A. | | 登録日 | 2021-06-01 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-05-31 | | 実験手法 | ELECTRON MICROSCOPY (3.33 Å) | | 主引用文献 | Inhibition mechanism of NKCC1 involves the carboxyl terminus and long-range conformational coupling.
Sci Adv, 8, 2022
|
|
7K8C
 
 | |
7K8A
 
 | |
7K8D
 
 | |
5DNY
 
 | | Structure of the ATPrS-Mre11/Rad50-DNA complex | | 分子名称: | DNA (27-MER), DNA double-strand break repair Rad50 ATPase,DNA double-strand break repair Rad50 ATPase, DNA double-strand break repair protein Mre11, ... | | 著者 | Liu, Y. | | 登録日 | 2015-09-10 | | 公開日 | 2016-02-24 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.11 Å) | | 主引用文献 | ATP-dependent DNA binding, unwinding, and resection by the Mre11/Rad50 complex.
Embo J., 35, 2016
|
|
7K8B
 
 | |
7K7M
 
 | | Crystal Structure of a membrane protein | | 分子名称: | Drug exporters of the RND superfamily-like protein, alpha-D-glucopyranose-(1-1)-6-O-decanoyl-alpha-D-glucopyranose | | 著者 | Su, C.-C. | | 登録日 | 2020-09-23 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.33 Å) | | 主引用文献 | Structures of the mycobacterial membrane protein MmpL3 reveal its mechanism of lipid transport.
Plos Biol., 19, 2021
|
|
1UKQ
 
 | | Crystal structure of cyclodextrin glucanotransferase complexed with a pseudo-maltotetraose derived from acarbose | | 分子名称: | 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, CALCIUM ION, Cyclomaltodextrin glucanotransferase, ... | | 著者 | Haga, K, Kanai, R, Sakamoto, O, Harata, K, Yamane, K. | | 登録日 | 2003-09-01 | | 公開日 | 2004-02-24 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Effects of Essential Carbohydrate/Aromatic Stacking Interaction with Tyr100 and Phe259 on Substrate Binding of Cyclodextrin Glycosyltransferase from Alkalophilic Bacillus sp. 1011
J.Biochem.(Tokyo), 134, 2003
|
|
1UKS
 
 | | Crystal structure of F183L/F259L mutant cyclodextrin glucanotransferase complexed with a pseudo-maltotetraose derived from acarbose | | 分子名称: | 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-beta-D-galactopyranose, 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, CALCIUM ION, ... | | 著者 | Haga, K, Kanai, R, Sakamoto, O, Harata, K, Yamane, K. | | 登録日 | 2003-09-01 | | 公開日 | 2004-02-24 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Effects of Essential Carbohydrate/Aromatic Stacking Interaction with Tyr100 and Phe259 on Substrate Binding of Cyclodextrin Glycosyltransferase from Alkalophilic Bacillus sp. 1011
J.Biochem.(Tokyo), 134, 2003
|
|
1UKT
 
 | | Crystal structure of Y100L mutant cyclodextrin glucanotransferase compexed with an acarbose | | 分子名称: | 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-galactopyranose, 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, CALCIUM ION, ... | | 著者 | Haga, K, Kanai, R, Sakamoto, O, Harata, K, Yamane, K. | | 登録日 | 2003-09-01 | | 公開日 | 2004-02-24 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Effects of Essential Carbohydrate/Aromatic Stacking Interaction with Tyr100 and Phe259 on Substrate Binding of Cyclodextrin Glycosyltransferase from Alkalophilic Bacillus sp. 1011
J.Biochem.(Tokyo), 134, 2003
|
|
4GTA
 
 | | T. Maritima FDTS with FAD, dUMP, and Folinic Acid | | 分子名称: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Mathews, I.I, Lesley, S.A, Kohen, A. | | 登録日 | 2012-08-28 | | 公開日 | 2012-10-17 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Folate binding site of flavin-dependent thymidylate synthase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3MS4
 
 | |
3MPB
 
 | |
4LYN
 
 | | Crystal structure of cyclin-dependent kinase 2 (cdk2-wt) complex with (2s)-n-(5-(((5-tert-butyl-1,3-oxazol-2-yl)methyl)sulfanyl)-1,3-thiazol-2-yl)-2-phenylpropanamide | | 分子名称: | (2S)-N-(5-{[(5-tert-butyl-1,3-oxazol-2-yl)methyl]sulfanyl}-1,3-thiazol-2-yl)-2-phenylpropanamide, Cyclin-dependent kinase 2 | | 著者 | Sack, J.S. | | 登録日 | 2013-07-31 | | 公開日 | 2013-10-02 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery of Aminothiazole Inhibitors of Cyclin-Dependent Kinase 2: Synthesis, X-Ray Crystallographic Analysis, and Biological Activities
J.Med.Chem., 45, 2002
|
|
4GTB
 
 | | T. Maritima FDTS with FAD, dUMP, and Raltitrexed. | | 分子名称: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Mathews, I.I, Lesley, S.A, Kohen, A. | | 登録日 | 2012-08-28 | | 公開日 | 2012-10-17 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Folate binding site of flavin-dependent thymidylate synthase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6JC2
 
 | | Crystal structure of the Fab fragment of ipilimumab | | 分子名称: | SULFATE ION, ipilimumab fab heavy chain, ipilimumab fab light chain | | 著者 | Heo, Y.S. | | 登録日 | 2019-01-27 | | 公開日 | 2019-09-11 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Crystal Structure of the Fab Fragment of an Anti-CTLA-4 Antibody, Ipilimumab, Used for Cancer Immunotherapy
Bull.Korean Chem.Soc., 40, 2019
|
|
