7KMZ
 
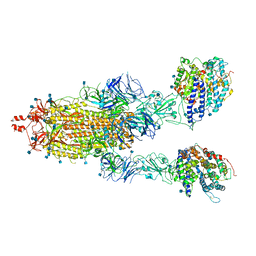 | | Cryo-EM structure of double ACE2-bound SARS-CoV-2 trimer Spike at pH 7.4 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Gorman, J, Kwong, P.D, Shapiro, L. | | 登録日 | 2020-11-03 | | 公開日 | 2020-12-09 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (3.62 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
6GBB
 
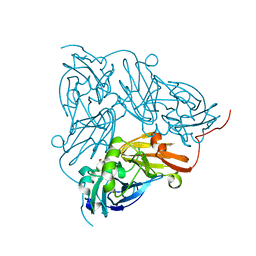 | | Copper nitrite reductase from Achromobacter cycloclastes: large cell polymorph dataset 1 | | 分子名称: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION | | 著者 | Ebrahim, A, Appleby, M.V, Axford, D, Beale, J, Moreno-Chicano, T, Sherrell, D.A, Strange, R.W, Owen, R.L, Hough, M.A. | | 登録日 | 2018-04-13 | | 公開日 | 2019-01-30 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Resolving polymorphs and radiation-driven effects in microcrystals using fixed-target serial synchrotron crystallography.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
7L61
 
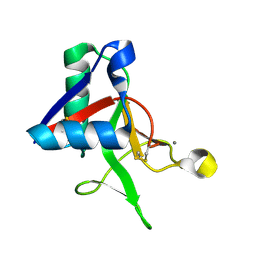 | |
7QU0
 
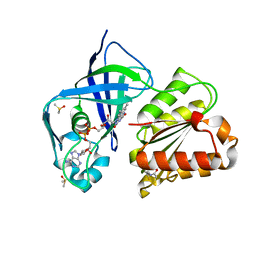 | | X-ray structure of FAD domain of NqrF of Klebsiella pneumoniae | | 分子名称: | DIMETHYL SULFOXIDE, FLAVIN-ADENINE DINUCLEOTIDE, Na(+)-translocating NADH-quinone reductase subunit F, ... | | 著者 | Stegmann, D, Steuber, J, Fritz, G. | | 登録日 | 2022-01-17 | | 公開日 | 2022-02-09 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Fast fragment- and compound-screening pipeline at the Swiss Light Source.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6GBY
 
 | | Copper nitrite reductase from Achromobacter cycloclastes: non-polymorph separated dataset 1 | | 分子名称: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION | | 著者 | Ebrahim, A, Appleby, M.V, Axford, D, Beale, J, Moreno-Chicano, T, Sherrell, D.A, Strange, R.W, Owen, R.L, Hough, M.A. | | 登録日 | 2018-04-16 | | 公開日 | 2019-01-30 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.48 Å) | | 主引用文献 | Resolving polymorphs and radiation-driven effects in microcrystals using fixed-target serial synchrotron crystallography.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
8FH5
 
 | |
8FH6
 
 | | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And Two AT-001 | | 分子名称: | (8-oxo-7-{[5-(trifluoromethyl)-1,3-benzothiazol-2-yl]methyl}-7,8-dihydropyrazino[2,3-d]pyridazin-5-yl)acetic acid, 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B1, ... | | 著者 | Arenas, R, Wilson, D.K. | | 登録日 | 2022-12-13 | | 公開日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.952 Å) | | 主引用文献 | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And Two AT-001
To Be Published
|
|
8FH7
 
 | |
8FH9
 
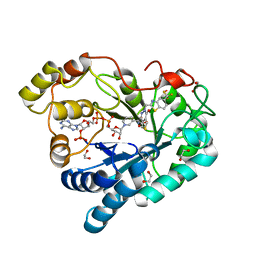 | | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And AT-007 | | 分子名称: | (4-oxo-3-{[5-(trifluoromethyl)-1,3-benzothiazol-2-yl]methyl}-3,4-dihydrothieno[3,4-d]pyridazin-1-yl)acetic acid, 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B1, ... | | 著者 | Arenas, R, Wilson, D.K. | | 登録日 | 2022-12-13 | | 公開日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And AT-007
To Be Published
|
|
8FH8
 
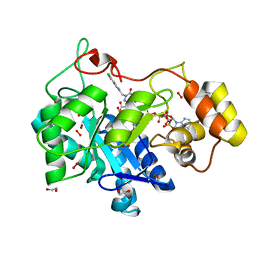 | |
7KJV
 
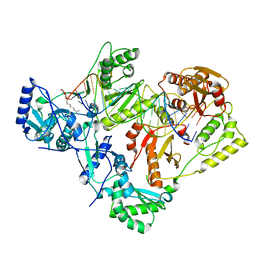 | | Structure of HIV-1 reverse transcriptase initiation complex core | | 分子名称: | HIV-1 viral RNA fragment, MAGNESIUM ION, Reverse transcriptase/ribonuclease H, ... | | 著者 | Ha, B, Larsen, K.P, Zhang, J, Fu, Z, Montabana, E, Jackson, L.N, Chen, D.H, Puglisi, E.V. | | 登録日 | 2020-10-26 | | 公開日 | 2021-03-17 | | 最終更新日 | 2021-05-12 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | High-resolution view of HIV-1 reverse transcriptase initiation complexes and inhibition by NNRTI drugs.
Nat Commun, 12, 2021
|
|
5IVX
 
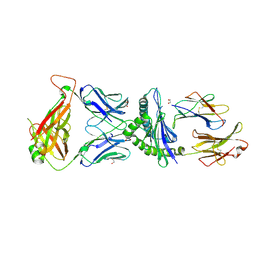 | | Crystal Structure of B4.2.3 T-Cell Receptor and H2-Dd P18-I10 Complex | | 分子名称: | 1,2-ETHANEDIOL, Beta-2-microglobulin, H-2 class I histocompatibility antigen, ... | | 著者 | Natarajan, K, Jiang, J, Margulies, D. | | 登録日 | 2016-03-21 | | 公開日 | 2017-03-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | An allosteric site in the T-cell receptor C beta domain plays a critical signalling role.
Nat Commun, 8, 2017
|
|
7WPO
 
 | | Structure of NeoCOV RBD binding to Bat37 ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Cao, L, Wang, X, Tortorici, M.A, Veesler, D. | | 登録日 | 2022-01-24 | | 公開日 | 2022-11-30 | | 最終更新日 | 2023-03-15 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Close relatives of MERS-CoV in bats use ACE2 as their functional receptors.
Nature, 612, 2022
|
|
7WPZ
 
 | | Structure of PDF-2180-COV RBD binding to Bat37 ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | 著者 | Cao, L, Wang, X, Tortorici, M.A, Veesler, D. | | 登録日 | 2022-01-24 | | 公開日 | 2022-11-30 | | 最終更新日 | 2023-03-15 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Close relatives of MERS-CoV in bats use ACE2 as their functional receptors.
Nature, 612, 2022
|
|
6STO
 
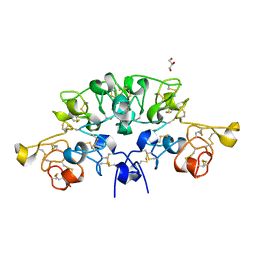 | | Three dimensional structure of the giant reed (Arundodonax) lectin (ADL) complex with N-Acetyl lactosamine | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Arundo donax Lectin (ADL), GLYCEROL, ... | | 著者 | Perduca, M, Monaco, H.L, Bovi, M, Destefanis, L, Nadali, D, Fin, L, Carrizo, M.E. | | 登録日 | 2019-09-11 | | 公開日 | 2021-07-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Three-dimensional structure and properties of the giant reed (Arundo donax) lectin (ADL).
Glycobiology, 2021
|
|
5LFN
 
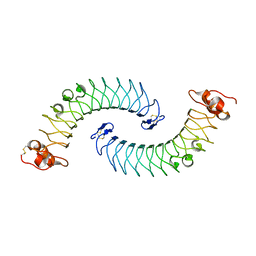 | |
6G8S
 
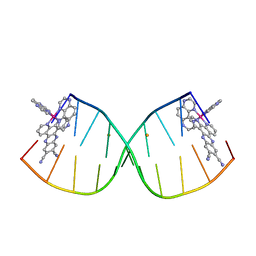 | | [Ru(TAP)2(11,12-CN2-dppz)]2+ bound to d(CCGGACCCGG/CCGGGTCCGG)2 | | 分子名称: | BARIUM ION, DNA (5'-D(*CP*CP*GP*GP*AP*CP*CP*CP*GP*G)-3'), DNA (5'-D(*CP*CP*GP*GP*GP*TP*CP*CP*GP*G)-3'), ... | | 著者 | McQuaid, K.T, Hall, J.P, Cardin, C.J. | | 登録日 | 2018-04-09 | | 公開日 | 2019-04-24 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.66 Å) | | 主引用文献 | X-ray Crystal Structures Show DNA Stacking Advantage of Terminal Nitrile Substitution in Ru-dppz Complexes.
Chemistry, 24, 2018
|
|
8CUL
 
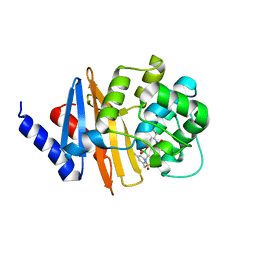 | | Xray ray crystal structure of OXA-24/40 in complex with CR167 | | 分子名称: | 3-({[(dihydroxyboranyl)methyl]sulfamoyl}methyl)benzoic acid, Beta-lactamase | | 著者 | Fernando, M.C, Wallar, B.J, Powers, R.A. | | 登録日 | 2022-05-17 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Sulfonamidoboronic Acids as "Cross-Class" Inhibitors of an Expanded-Spectrum Class C Cephalosporinase, ADC-33, and a Class D Carbapenemase, OXA-24/40: Strategic Compound Design to Combat Resistance in Acinetobacter baumannii .
Antibiotics, 12, 2023
|
|
8CUO
 
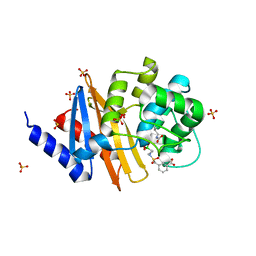 | | X-ray crystal structure of OXA-24/40 in complex with sulfonamidoboronic acid 6e | | 分子名称: | 3-({[(1R)-1-boronopropyl]sulfamoyl}methyl)benzoic acid, Beta-lactamase, SULFATE ION | | 著者 | Fernando, M.C, Wallar, B.J, Powers, R.A. | | 登録日 | 2022-05-17 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Sulfonamidoboronic Acids as "Cross-Class" Inhibitors of an Expanded-Spectrum Class C Cephalosporinase, ADC-33, and a Class D Carbapenemase, OXA-24/40: Strategic Compound Design to Combat Resistance in Acinetobacter baumannii .
Antibiotics, 12, 2023
|
|
8CUM
 
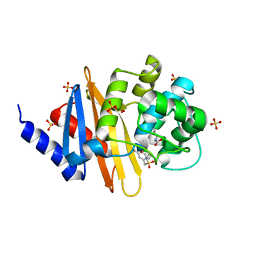 | | X-ray crystal structure of OXA-24/40 in complex with sulfonamidoboronic acid 6d | | 分子名称: | 3-({[(1S)-1-boronopropyl]sulfamoyl}methyl)benzoic acid, Beta-lactamase, SULFATE ION | | 著者 | Fernando, M.C, Wallar, B.J, Powers, R.A. | | 登録日 | 2022-05-17 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Sulfonamidoboronic Acids as "Cross-Class" Inhibitors of an Expanded-Spectrum Class C Cephalosporinase, ADC-33, and a Class D Carbapenemase, OXA-24/40: Strategic Compound Design to Combat Resistance in Acinetobacter baumannii .
Antibiotics, 12, 2023
|
|
6T8F
 
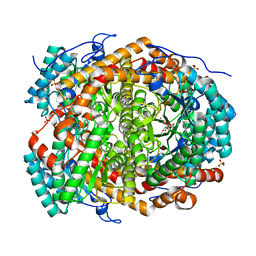 | |
6TN1
 
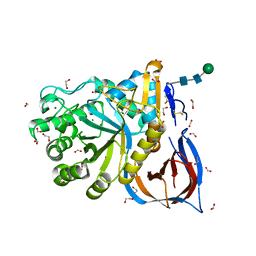 | | Unliganded Crystal Structure of Recombinant GBA | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, FORMIC ACID, ... | | 著者 | Rowland, R.J, Davies, G.J. | | 登録日 | 2019-12-05 | | 公開日 | 2020-06-10 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (0.98 Å) | | 主引用文献 | A baculoviral system for the production of human beta-glucocerebrosidase enables atomic resolution analysis.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6GCG
 
 | | Copper nitrite reductase from Achromobacter cycloclastes: large polymorph dataset 15 | | 分子名称: | COPPER (II) ION, Copper-containing nitrite reductase | | 著者 | Ebrahim, A, Appleby, M.V, Axford, D, Beale, J, Moreno-Chicano, T, Sherrell, D.A, Strange, R.W, Owen, R.L, Hough, M.A. | | 登録日 | 2018-04-17 | | 公開日 | 2019-01-30 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.80015242 Å) | | 主引用文献 | Resolving polymorphs and radiation-driven effects in microcrystals using fixed-target serial synchrotron crystallography.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6PUL
 
 | | Structure of human MAIT A-F7 TCR in complex with human MR1 3'D-5-OP-RU | | 分子名称: | 1,3-dideoxy-1-({2,6-dioxo-5-[(E)-(2-oxopropylidene)amino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-erythro-pentitol, ACETATE ION, Beta-2-microglobulin, ... | | 著者 | Awad, W, Keller, A.N, Rossjohn, J. | | 登録日 | 2019-07-18 | | 公開日 | 2020-02-19 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | The molecular basis underpinning the potency and specificity of MAIT cell antigens.
Nat.Immunol., 21, 2020
|
|
6GF0
 
 | | Lysozyme structure determined from SFX data using a Sheet-on-Sheet chipless chip | | 分子名称: | Lysozyme C | | 著者 | Doak, R.B, Gorel, A, Foucar, L, Gruenbein, M.L, Hilpert, M, Kloos, M, Nass Kovacs, G, Roome, C, Shoeman, R.L, Stricker, M, Tono, K, You, D, Ueda, K, Sherrel, D, Owen, R, Barends, T.R.M, Schlichting, I. | | 登録日 | 2018-04-27 | | 公開日 | 2018-10-17 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Crystallography on a chip - without the chip: sheet-on-sheet sandwich.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
