8UAO
 
 | |
8UBG
 
 | | DpHF19 filament | | 分子名称: | DpHF19,Green fluorescent protein (Fragment) | | 著者 | Lynch, E.M, Shen, H, Kollman, J.M, Baker, D. | | 登録日 | 2023-09-22 | | 公開日 | 2024-04-10 | | 最終更新日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | De novo design of pH-responsive self-assembling helical protein filaments.
Nat Nanotechnol, 19, 2024
|
|
8UHW
 
 | |
8VCT
 
 | |
8VCJ
 
 | |
1MGV
 
 | | Crystal Structure of the R391A Mutant of 7,8-Diaminopelargonic Acid Synthase | | 分子名称: | 7,8-diamino-pelargonic acid aminotransferase, ISOPROPYL ALCOHOL, PYRIDOXAL-5'-PHOSPHATE, ... | | 著者 | Eliot, A.C, Sandmark, J, Schneider, G, Kirsch, J.F. | | 登録日 | 2002-08-16 | | 公開日 | 2002-11-27 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The Dual-Specific Active Site of 7,8-Diaminopelargonic Acid Synthase and the Effect of the R391A Mutation
Biochemistry, 41, 2002
|
|
1PEZ
 
 | | Bacillus circulans strain 251 mutant A230V | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETIC ACID, ... | | 著者 | Rozeboom, H.J, Dijkstra, B.W. | | 登録日 | 2003-05-23 | | 公開日 | 2003-10-28 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.32 Å) | | 主引用文献 | Conversion of Cyclodextrin Glycosyltransferase into a Starch Hydrolase by Directed Evolution: The Role of Alanine 230 in Acceptor Subsite +1
Biochemistry, 42, 2003
|
|
2W5B
 
 | | Human Nek2 kinase ATPgammaS-bound | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | 著者 | Bayliss, R. | | 登録日 | 2008-12-08 | | 公開日 | 2008-12-23 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Insights Into the Conformational Variability and Regulation of Human Nek2 Kinase.
J.Mol.Biol., 386, 2009
|
|
1XYA
 
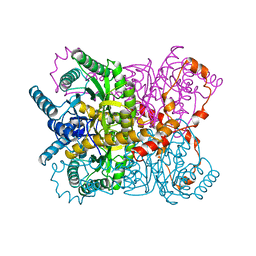 | | X-RAY CRYSTALLOGRAPHIC STRUCTURES OF D-XYLOSE ISOMERASE-SUBSTRATE COMPLEXES POSITION THE SUBSTRATE AND PROVIDE EVIDENCE FOR METAL MOVEMENT DURING CATALYSIS | | 分子名称: | HYDROXIDE ION, MAGNESIUM ION, XYLOSE ISOMERASE | | 著者 | Lavie, A, Allen, K.N, Petsko, G.A, Ringe, D. | | 登録日 | 1994-01-03 | | 公開日 | 1994-05-31 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | X-ray crystallographic structures of D-xylose isomerase-substrate complexes position the substrate and provide evidence for metal movement during catalysis.
Biochemistry, 33, 1994
|
|
1XYC
 
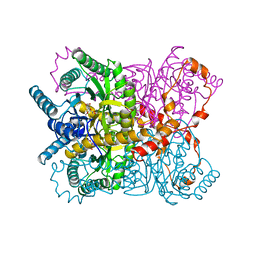 | | X-RAY CRYSTALLOGRAPHIC STRUCTURES OF D-XYLOSE ISOMERASE-SUBSTRATE COMPLEXES POSITION THE SUBSTRATE AND PROVIDE EVIDENCE FOR METAL MOVEMENT DURING CATALYSIS | | 分子名称: | 3-O-METHYLFRUCTOSE IN LINEAR FORM, MAGNESIUM ION, XYLOSE ISOMERASE | | 著者 | Lavie, A, Allen, K.N, Petsko, G.A, Ringe, D. | | 登録日 | 1994-01-03 | | 公開日 | 1994-05-31 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.19 Å) | | 主引用文献 | X-ray crystallographic structures of D-xylose isomerase-substrate complexes position the substrate and provide evidence for metal movement during catalysis.
Biochemistry, 33, 1994
|
|
1XYB
 
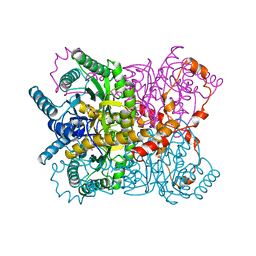 | | X-RAY CRYSTALLOGRAPHIC STRUCTURES OF D-XYLOSE ISOMERASE-SUBSTRATE COMPLEXES POSITION THE SUBSTRATE AND PROVIDE EVIDENCE FOR METAL MOVEMENT DURING CATALYSIS | | 分子名称: | D-glucose, MAGNESIUM ION, XYLOSE ISOMERASE | | 著者 | Lavie, A, Allen, K.N, Petsko, G.A, Ringe, D. | | 登録日 | 1994-01-03 | | 公開日 | 1994-05-31 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | X-ray crystallographic structures of D-xylose isomerase-substrate complexes position the substrate and provide evidence for metal movement during catalysis.
Biochemistry, 33, 1994
|
|
2G2F
 
 | |
152L
 
 | |
1JMF
 
 | |
1JMH
 
 | |
1JMI
 
 | |
1JMG
 
 | |
8UQE
 
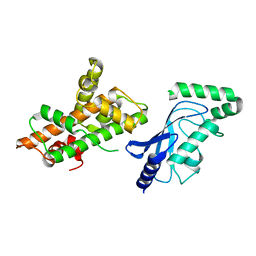 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 26-residue linker (RING not modeled in density) | | 分子名称: | E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E | | 著者 | Hu, Q, Botuyan, M.V, Mer, G. | | 登録日 | 2023-10-23 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.562 Å) | | 主引用文献 | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQD
 
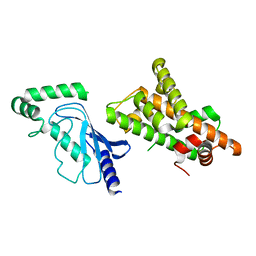 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 20-residue linker (condition 2. RING not modeled in density) | | 分子名称: | E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E | | 著者 | Hu, Q, Botuyan, M.V, Mer, G. | | 登録日 | 2023-10-23 | | 公開日 | 2024-01-17 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.893 Å) | | 主引用文献 | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
2CEP
 
 | |
1E0D
 
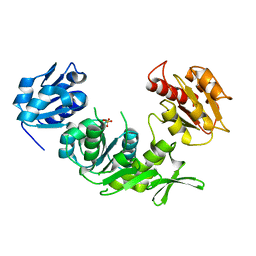 | | UDP-N-Acetylmuramoyl-L-Alanine:D-Glutamate Ligase | | 分子名称: | SULFATE ION, UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE | | 著者 | Fanchon, E, Bertrand, J, Chantalat, L, Dideberg, O. | | 登録日 | 2000-03-24 | | 公開日 | 2000-06-09 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | "Open" Structures of Murd: Domain Movements and Structural Similarities with Folylpolyglutamate Synthetase.
J.Mol.Biol., 301, 2000
|
|
2LT9
 
 | |
7ASQ
 
 | |
7AM9
 
 | | OMPD-domain of human UMPS in complex with the substrate OMP at 0.99 Angstroms resolution | | 分子名称: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | 著者 | Tittmann, K, Rindfleisch, S, Krull, M. | | 登録日 | 2020-10-08 | | 公開日 | 2021-11-03 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (0.99 Å) | | 主引用文献 | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
7EDD
 
 | |
