7K80
 
 | | KIR3DL1*001 in complex with HLA-A*24:02 presenting the RYPLTFGW peptide | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ARG-TYR-PRO-LEU-THR-PHE-GLY-TRP, ... | | 著者 | MacLachlan, B.J, Rossjohn, J, Vivian, J.P. | | 登録日 | 2020-09-24 | | 公開日 | 2020-12-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The Role of the HLA Class I alpha 2 Helix in Determining Ligand Hierarchy for the Killer Cell Ig-like Receptor 3DL1.
J Immunol., 206, 2021
|
|
6HX1
 
 | | IRE1 ALPHA IN COMPLEX WITH imidazo[1,2-b]pyridazin-8-amine compound 2 | | 分子名称: | 6-chloranyl-~{N}-(cyclopropylmethyl)-3-(2~{H}-indazol-5-yl)imidazo[1,2-b]pyridazin-8-amine, Serine/threonine-protein kinase/endoribonuclease IRE1 | | 著者 | Augustin, M.A, Krapp, S, Bayliss, R, Collins, I. | | 登録日 | 2018-10-15 | | 公開日 | 2019-02-27 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Binding to an Unusual Inactive Kinase Conformation by Highly Selective Inhibitors of Inositol-Requiring Enzyme 1 alpha Kinase-Endoribonuclease.
J.Med.Chem., 62, 2019
|
|
5ZMC
 
 | | Structural Basis for Reactivation of -146C>T Mutant TERT Promoter by cooperative binding of p52 and ETS1/2 | | 分子名称: | DNA (5'-D(P*CP*GP*GP*GP*GP*AP*CP*CP*CP*GP*GP*AP*AP*GP*GP*G)-3'), DNA (5'-D(P*GP*CP*CP*CP*TP*TP*CP*CP*GP*GP*GP*TP*CP*CP*CP*C)-3'), Nuclear factor NF-kappa-B p100 subunit, ... | | 著者 | Xu, X, Bharath, S.R, Song, H. | | 登録日 | 2018-04-02 | | 公開日 | 2018-09-19 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.99 Å) | | 主引用文献 | Structural basis for reactivating the mutant TERT promoter by cooperative binding of p52 and ETS1.
Nat Commun, 9, 2018
|
|
8X7J
 
 | | Cryo-EM structures of RNF168/UbcH5c-Ub/nucleosomes complex determined by activity-based chemical trapping strategy | | 分子名称: | DNA (144-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | 著者 | Ai, H.S, Tong, Z.B, Deng, Z.H, Pan, M, Liu, L. | | 登録日 | 2023-11-24 | | 公開日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (3.39 Å) | | 主引用文献 | Capturing Snapshots of Nucleosomal H2A K13/K15 Ubiquitination Mediated by the Monomeric E3 Ligase RNF168
Biorxiv, 2024
|
|
8X7K
 
 | | Cryo-EM structures of RNF168/UbcH5c-Ub in complex with H2AK13Ub nucleosomes determined by activity-based chemical trapping strategy (adjacent H2AK13/15 dual-monoubiquitination) | | 分子名称: | DNA (143-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | 著者 | Ai, H.S, Tong, Z.B, Deng, Z.H, Pan, M, Liu, L. | | 登録日 | 2023-11-24 | | 公開日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (3.27 Å) | | 主引用文献 | Capturing Snapshots of Nucleosomal H2A K13/K15 Ubiquitination Mediated by the Monomeric E3 Ligase RNF168
Biorxiv, 2024
|
|
8X7I
 
 | | Cryo-EM structures of RNF168/UbcH5c-Ub in complex with H2AK13Ub nucleosomes determined by intein-based E2-Ub-NCP conjugation strategy | | 分子名称: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | 著者 | Ai, H.S, Tong, Z.B, Deng, Z.H, Pan, M, Liu, L. | | 登録日 | 2023-11-24 | | 公開日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (3.27 Å) | | 主引用文献 | Capturing Snapshots of Nucleosomal H2A K13/K15 Ubiquitination Mediated by the Monomeric E3 Ligase RNF168
Biorxiv, 2024
|
|
6FNL
 
 | | Crystal Structure of Ephrin B4 (EphB4) Receptor Protein Kinase | | 分子名称: | Ephrin type-B receptor 4 | | 著者 | Kudlinzki, D, Troester, A, Witt, K, Linhard, V.L, Saxena, K, Schwalbe, H. | | 登録日 | 2018-02-04 | | 公開日 | 2018-08-08 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.269 Å) | | 主引用文献 | NVP-BHG712: Effects of Regioisomers on the Affinity and Selectivity toward the EPHrin Family.
ChemMedChem, 13, 2018
|
|
6S6B
 
 | | Type III-B Cmr-beta Cryo-EM structure of the Apo state | | 分子名称: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | 著者 | Sofos, N, Montoya, G, Stella, S. | | 登録日 | 2019-07-02 | | 公開日 | 2020-07-08 | | 最終更新日 | 2020-09-16 | | 実験手法 | ELECTRON MICROSCOPY (2.75 Å) | | 主引用文献 | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
6FNI
 
 | | Crystal Structure of Ephrin B4 (EphB4) Receptor Protein Kinase with NVP-BHG712 | | 分子名称: | 4-methyl-3-[(1-methyl-6-pyridin-3-yl-pyrazolo[3,4-d]pyrimidin-4-yl)amino]-~{N}-[3-(trifluoromethyl)phenyl]benzamide, Ephrin type-B receptor 4 | | 著者 | Kudlinzki, D, Troester, A, Witt, K, Linhard, V.L, Saxena, K, Schwalbe, H. | | 登録日 | 2018-02-04 | | 公開日 | 2018-08-08 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.468 Å) | | 主引用文献 | NVP-BHG712: Effects of Regioisomers on the Affinity and Selectivity toward the EPHrin Family.
ChemMedChem, 13, 2018
|
|
8U5M
 
 | | Structure of Sts-1 HP domain with rebamipide | | 分子名称: | Rebamipide, Ubiquitin-associated and SH3 domain-containing protein B | | 著者 | Azia, F, Dey, R, French, J.B. | | 登録日 | 2023-09-12 | | 公開日 | 2024-02-07 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | Rebamipide and Derivatives are Potent, Selective Inhibitors of Histidine Phosphatase Activity of the Suppressor of T Cell Receptor Signaling Proteins.
J.Med.Chem., 67, 2024
|
|
6QKI
 
 | | Native structure of EgtB from Chloracidobacterium thermophilum, a type II sulfoxide synthase | | 分子名称: | FE (III) ION, Uncharacterized protein | | 著者 | Stampfli, A.R, Badri, B.N, Schirmer, T, Seebeck, F.P. | | 登録日 | 2019-01-29 | | 公開日 | 2019-03-27 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | An Alternative Active Site Architecture for O2Activation in the Ergothioneine Biosynthetic EgtB from Chloracidobacterium thermophilum.
J.Am.Chem.Soc., 141, 2019
|
|
7KFK
 
 | |
4R9K
 
 | | Structure of thermostable eightfold mutant of limonene epoxide hydrolase from Rhodococcus erythropolis | | 分子名称: | (2R)-2-hydroxyhexanamide, GLYCEROL, Limonene-1,2-epoxide hydrolase | | 著者 | Floor, R.J, Wijma, H.J, Jekel, P.A, Terwisscha van Scheltinga, A.C, Dijkstra, B.W, Janssen, D.B. | | 登録日 | 2014-09-05 | | 公開日 | 2014-09-24 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | X-ray crystallographic validation of structure predictions used in computational design for protein stabilization.
Proteins, 83, 2015
|
|
7EDH
 
 | | Cryo-EM structure of SARS-CoV-2 S-UK variant (B.1.1.7), one RBD-up conformation 3 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Yang, T.J, Yu, P.Y, Chang, Y.C, Wu, H.C, Hsu, S.T.D. | | 登録日 | 2021-03-16 | | 公開日 | 2021-09-01 | | 最終更新日 | 2022-01-05 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Effect of SARS-CoV-2 B.1.1.7 mutations on spike protein structure and function.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7EDF
 
 | | Cryo-EM structure of SARS-CoV-2 S-UK variant (B.1.1.7), one RBD-up conformation 1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Yang, T.J, Yu, P.Y, Chang, Y.C, Wu, H.C, Hsu, S.T.D. | | 登録日 | 2021-03-16 | | 公開日 | 2021-09-01 | | 最終更新日 | 2022-01-05 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Effect of SARS-CoV-2 B.1.1.7 mutations on spike protein structure and function.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7EDG
 
 | | Cryo-EM structure of SARS-CoV-2 S-UK variant (B.1.1.7), one RBD-up conformation 2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Yang, T.J, Yu, P.Y, Chang, Y.C, Wu, H.C, Hsu, S.T.D. | | 登録日 | 2021-03-16 | | 公開日 | 2021-09-01 | | 最終更新日 | 2022-01-05 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Effect of SARS-CoV-2 B.1.1.7 mutations on spike protein structure and function.
Nat.Struct.Mol.Biol., 28, 2021
|
|
3HO6
 
 | |
4XAB
 
 | | Crystal Structure of EvdO2 from Micromonospora carbonacea var. aurantiaca | | 分子名称: | EvdO2, IMIDAZOLE, NICKEL (II) ION | | 著者 | McCulloch, K.M, McCranie, E.K, Sarwar, M, Mathieu, J.L, Gitschlag, B.L, Du, Y, Bachmann, B.O, Iverson, T.M. | | 登録日 | 2014-12-13 | | 公開日 | 2015-08-05 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Oxidative cyclizations in orthosomycin biosynthesis expand the known chemistry of an oxygenase superfamily.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XAA
 
 | | Crystal Structure of AviO1 from Streptomyces viridochromogenes Tue57 | | 分子名称: | NICKEL (II) ION, Putative oxygenase | | 著者 | McCulloch, K.M, McCranie, E.K, Sarwar, M, Mathieu, J.L, Gitschlag, B.L, Du, Y, Bachmann, B.O, Iverson, T.M. | | 登録日 | 2014-12-13 | | 公開日 | 2015-08-05 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Oxidative cyclizations in orthosomycin biosynthesis expand the known chemistry of an oxygenase superfamily.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6S8B
 
 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA and AMPPnP, state 1 | | 分子名称: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | 著者 | Sofos, N, Montoya, G, Stella, S. | | 登録日 | 2019-07-09 | | 公開日 | 2020-07-08 | | 最終更新日 | 2020-09-16 | | 実験手法 | ELECTRON MICROSCOPY (2.41 Å) | | 主引用文献 | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
6S91
 
 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA and AMPPnP, state 2 | | 分子名称: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | 著者 | Sofos, N, Montoya, G, Stella, S. | | 登録日 | 2019-07-11 | | 公開日 | 2020-07-08 | | 最終更新日 | 2020-09-16 | | 実験手法 | ELECTRON MICROSCOPY (2.68 Å) | | 主引用文献 | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
6FNM
 
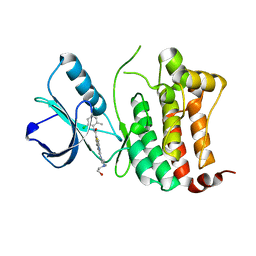 | | Crystal Structure of Ephrin B4 (EphB4) Receptor Protein Kinase with Dasatinib | | 分子名称: | Ephrin type-B receptor 4, N-(2-CHLORO-6-METHYLPHENYL)-2-({6-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]-2-METHYLPYRIMIDIN-4-YL}AMINO)-1,3-THIAZOLE-5-CARBOXAMIDE | | 著者 | Kudlinzki, D, Troester, A, Witt, K, Linhard, V.L, Saxena, K, Schwalbe, H. | | 登録日 | 2018-02-04 | | 公開日 | 2018-08-08 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.157 Å) | | 主引用文献 | NVP-BHG712: Effects of Regioisomers on the Affinity and Selectivity toward the EPHrin Family.
ChemMedChem, 13, 2018
|
|
6FM4
 
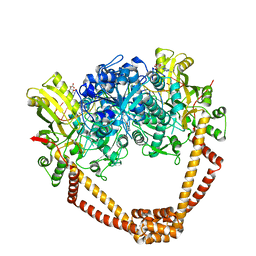 | | The crystal structure of S. aureus Gyrase complex with ID-130 and DNA | | 分子名称: | 5'-O-CARBOXY-2'-DEOXYADENOSINE, DNA (5'-5UA*D(P*GP*CP*CP*GP*TP*AP*GP*GP*GP*CP*CP*CP*TP*AP*CP*GP*GP*C)-3'), DNA (5'-5UA*D(P*GP*CP*CP*GP*TP*AP*GP*GP*GP*CP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | 著者 | Ombrato, R, Garofalo, B, Mangano, G, Mancini, F. | | 登録日 | 2018-01-30 | | 公開日 | 2019-07-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Virtual Screening Approach and Investigation of Structure-Activity Relationships To Discover Novel Bacterial Topoisomerase Inhibitors Targeting Gram-Positive and Gram-Negative Pathogens.
J.Med.Chem., 62, 2019
|
|
6C6Y
 
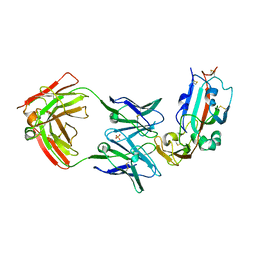 | | Crystal structure of Middle-East Respiratory Syndrome (MERS) coronavirus neutralizing antibody JC57-14 isolated from a vaccinated rhesus macaque in complex with MERS Receptor Binding Domain | | 分子名称: | JC57-14 Heavy chain, JC57-14 Light chain, SULFATE ION, ... | | 著者 | Joyce, M.G, Mascola, J.R, Graham, B.S, Kwong, P.D. | | 登録日 | 2018-01-19 | | 公開日 | 2018-03-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.32 Å) | | 主引用文献 | Importance of Neutralizing Monoclonal Antibodies Targeting Multiple Antigenic Sites on the Middle East Respiratory Syndrome Coronavirus Spike Glycoprotein To Avoid Neutralization Escape.
J. Virol., 92, 2018
|
|
6SIC
 
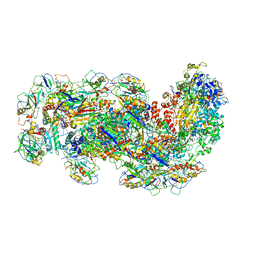 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA | | 分子名称: | CRISPR-associated RAMP protein, Cmr1 family, Cmr4 family, ... | | 著者 | Sofos, N, Montoya, G, Stella, S. | | 登録日 | 2019-08-09 | | 公開日 | 2020-07-08 | | 最終更新日 | 2020-09-16 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
