1Y82
 
 | | Conserved hypothetical protein Pfu-367848-001 from Pyrococcus furiosus | | 分子名称: | UNKNOWN ATOM OR ION, hypothetical protein | | 著者 | Horanyi, P, Tempel, W, Habel, J, Chen, L, Lee, D, Nguyen, D, Chang, S.-H, Florence, Q, Zhou, W, Lin, D, Zhang, H, Praissman, J, Jenney Jr, F.E, Adams, M.W.W, Liu, Z.-J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | 登録日 | 2004-12-10 | | 公開日 | 2005-01-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Conserved hypothetical protein Pfu-367848-001 from Pyrococcus furiosus
To be published
|
|
1M6G
 
 | | Structural Characterisation of the Holliday Junction TCGGTACCGA | | 分子名称: | 5'-D(*TP*CP*GP*GP*TP*AP*CP*CP*GP*A)-3', STRONTIUM ION | | 著者 | Thorpe, J.H, Gale, B.C, Teixeira, S.C.M, Cardin, C.J. | | 登録日 | 2002-07-16 | | 公開日 | 2003-05-06 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.652 Å) | | 主引用文献 | Conformational and hydration effects of site-selective sodium, calcium and
strontium ion binding to the DNA Holliday junction structure
d(TCGGTACCGA)(4)
J.Mol.Biol., 327, 2003
|
|
7MMR
 
 | |
7MPJ
 
 | | Stm1 bound vacant 80S structure isolated from wild-type | | 分子名称: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | 著者 | Rai, J, Zhao, Y, Li, H. | | 登録日 | 2021-05-04 | | 公開日 | 2022-05-11 | | 最終更新日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | CryoEM structures of pseudouridine-free ribosome suggest impacts of chemical modifications on ribosome conformations.
Structure, 30, 2022
|
|
7MPI
 
 | | Stm1 bound vacant 80S structure isolated from cbf5-D95A | | 分子名称: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | 著者 | Rai, J, Zhao, Y, Li, H. | | 登録日 | 2021-05-04 | | 公開日 | 2022-05-11 | | 最終更新日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | CryoEM structures of pseudouridine-free ribosome suggest impacts of chemical modifications on ribosome conformations.
Structure, 30, 2022
|
|
7M5D
 
 | |
7M7X
 
 | |
7NEF
 
 | | Fucosylated linear peptide Fln65 bound to the fucose binding lectin LecB PA-IIL from Pseudomonas aeruginosa at 1.5 Angstrom resolution | | 分子名称: | 3,7-anhydro-2,8-dideoxy-L-glycero-D-gluco-octonic acid, CALCIUM ION, Fln65, ... | | 著者 | Personne, H, Baeriswyl, S, Stocker, A, Reymond, J.-L. | | 登録日 | 2021-02-03 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.51 Å) | | 主引用文献 | A mixed chirality alpha-helix in a stapled bicyclic and a linear antimicrobial peptide revealed by X-ray crystallography.
Rsc Chem Biol, 2, 2021
|
|
7ND4
 
 | | EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-88 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-88 Fab heavy chain, ... | | 著者 | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | 登録日 | 2021-01-30 | | 公開日 | 2021-03-03 | | 最終更新日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7ND3
 
 | | EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-40 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-40 heavy chain, ... | | 著者 | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | 登録日 | 2021-01-30 | | 公開日 | 2021-03-03 | | 最終更新日 | 2023-11-29 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7ND7
 
 | | EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-316 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | 登録日 | 2021-01-30 | | 公開日 | 2021-03-03 | | 最終更新日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7ND8
 
 | | EM structure of SARS-CoV-2 Spike glycoprotein in complex with COVOX-384 Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-384 Fab heavy chain, ... | | 著者 | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | 登録日 | 2021-01-30 | | 公開日 | 2021-03-03 | | 最終更新日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7NDC
 
 | | EM structure of SARS-CoV-2 Spike glycoprotein (all RBD down) in complex with COVOX-159 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-159 Fab light chain, ... | | 著者 | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | 登録日 | 2021-01-30 | | 公開日 | 2021-03-03 | | 最終更新日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (4.1 Å) | | 主引用文献 | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
7ND9
 
 | | EM structure of SARS-CoV-2 Spike glycoprotein (one RBD up) in complex with COVOX-253H55L Fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Duyvesteyn, H.M.E, Zhao, Y, Ren, J, Stuart, D. | | 登録日 | 2021-01-30 | | 公開日 | 2021-03-03 | | 最終更新日 | 2021-04-28 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | The antigenic anatomy of SARS-CoV-2 receptor binding domain.
Cell, 184, 2021
|
|
1YSY
 
 | | NMR Structure of the nonstructural Protein 7 (nsP7) from the SARS CoronaVirus | | 分子名称: | Replicase polyprotein 1ab (pp1ab) (ORF1AB) | | 著者 | Peti, W, Herrmann, T, Johnson, M.A, Kuhn, P, Stevens, R.C, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | 登録日 | 2005-02-09 | | 公開日 | 2005-12-06 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural genomics of the severe acute respiratory syndrome coronavirus: nuclear magnetic resonance structure of the protein nsP7.
J.Virol., 79, 2005
|
|
1OS9
 
 | | Binary enzyme-product complexes of human MMP12 | | 分子名称: | CALCIUM ION, Macrophage metalloelastase, ZINC ION | | 著者 | Bertini, I, Calderone, V, Fragai, M, Luchinat, C, Mangani, S, Terni, B. | | 登録日 | 2003-03-19 | | 公開日 | 2003-08-05 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | X-ray Structures of Binary and Ternary Enzyme-Product-Inhibitor Complexes of Matrix Metalloproteinases
Angew.Chem.Int.Ed.Engl., 42, 2003
|
|
1P8J
 
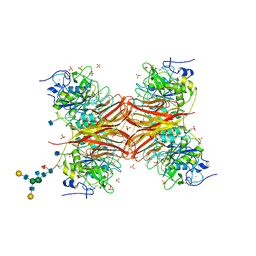 | | CRYSTAL STRUCTURE OF THE PROPROTEIN CONVERTASE FURIN | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, DECANOYL-ARG-VAL-LYS-ARG-CHLOROMETHYLKETONE INHIBITOR, ... | | 著者 | Henrich, S, Cameron, A, Bourenkov, G.P, Kiefersauer, R, Huber, R, Lindberg, I, Bode, W, Than, M.E. | | 登録日 | 2003-05-07 | | 公開日 | 2003-07-08 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The Crystal Structure of the Proprotein Processing Proteinase Furin Explains its Stringent Specificity
Nat.Struct.Biol., 10, 2003
|
|
4IN5
 
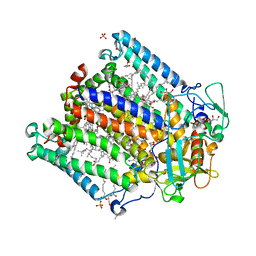 | | (M)L214G mutant of the Rhodobacter sphaeroides Reaction Center | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | 著者 | Saer, R.G, Hardjasa, A, Murphy, M.E, Beatty, J.T. | | 登録日 | 2013-01-04 | | 公開日 | 2013-05-01 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Role of Rhodobacter sphaeroides Photosynthetic Reaction Center Residue M214 in the Composition, Absorbance Properties, and Conformations of HA and BA Cofactors.
Biochemistry, 52, 2013
|
|
4IGG
 
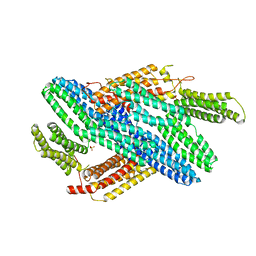 | |
4IL6
 
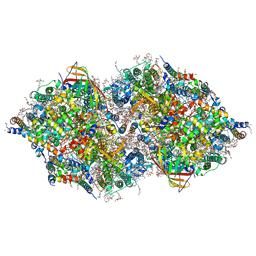 | | Structure of Sr-substituted photosystem II | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Koua, F.H.M, Umena, Y, Kawakami, K, Kamiya, N, Shen, J.R. | | 登録日 | 2012-12-29 | | 公開日 | 2013-03-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of Sr-substituted photosystem II at 2.1 A resolution and its implications in the mechanism of water oxidation
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4IUJ
 
 | |
4IYM
 
 | | Crystal structure of putative methylmalonate-semialdehyde dehydrogenase from Sinorhizobium meliloti 1021 complexed with NAD, target 011934 | | 分子名称: | MAGNESIUM ION, Methylmalonate-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2013-01-28 | | 公開日 | 2013-04-03 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of putative methylmalonate-semialdehyde dehydrogenase from Sinorhizobium meliloti 1021 complexed with NAD, target 011934
To be Published
|
|
4IXR
 
 | | RT fs X-ray diffraction of Photosystem II, first illuminated state | | 分子名称: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | 著者 | Kern, J, Alonso-Mori, R, Tran, R, Hattne, J, Gildea, R.J, Echols, N, Gloeckner, C, Hellmich, J, Laksmono, H, Sierra, R.G, Lassalle-Kaiser, B, Koroidov, S, Lampe, A, Han, G, Gul, S, DiFiore, D, Milathianaki, D, Fry, A.R, Miahnahri, A, Schafer, D.W, Messerschmidt, M, Seibert, M.M, Koglin, J.E, Sokaras, D, Weng, T.-C, Sellberg, J, Latimer, M.J, Grosse-Kunstleve, R.W, Zwart, P.H, White, W.E, Glatzel, P, Adams, P.D, Bogan, M.J, Williams, G.J, Boutet, S, Messinger, J, Zouni, A, Sauter, N.K, Yachandra, V.K, Bergmann, U, Yano, J. | | 登録日 | 2013-01-27 | | 公開日 | 2013-02-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (5.9 Å) | | 主引用文献 | Simultaneous femtosecond X-ray spectroscopy and diffraction of photosystem II at room temperature.
Science, 340, 2013
|
|
4IRE
 
 | | Crystal structure of GLIC with mutations at the loop C region | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ACETATE ION, OXALATE ION, ... | | 著者 | Chen, Q, Pan, J, Liang, Y.H, Xu, Y, Tang, P. | | 登録日 | 2013-01-14 | | 公開日 | 2013-04-24 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.19 Å) | | 主引用文献 | Signal transduction pathways in the pentameric ligand-gated ion channels.
Plos One, 8, 2013
|
|
4ITA
 
 | | Structure of bacterial enzyme in complex with cofactor | | 分子名称: | 1,2-ETHANEDIOL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Succinate-semialdehyde dehydrogenase | | 著者 | Rhee, S, Park, J. | | 登録日 | 2013-01-18 | | 公開日 | 2013-04-24 | | 最終更新日 | 2013-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structural Basis for a Cofactor-dependent Oxidation Protection and Catalysis of Cyanobacterial Succinic Semialdehyde Dehydrogenase.
J.Biol.Chem., 288, 2013
|
|
