7K51
 
 | | Mid-translocated non-frameshifting(CCA-A) complex with EF-G and GDPCP (Structure II) | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | 登録日 | 2020-09-16 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
7K50
 
 | | Pre-translocation non-frameshifting(CCA-A) complex (Structure I) | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | 登録日 | 2020-09-16 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
7K54
 
 | | Mid-translocated +1-frameshifting(CCC-A) complex with EF-G and GDPCP (Structure II-FS) | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | 登録日 | 2020-09-16 | | 公開日 | 2021-07-28 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
7L8F
 
 | | BG505 SOSIP.v5.2(7S) in complex with the polyclonal Fab pAbC-2 from animal Rh.33172 (Wk38 time point) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | 著者 | Antanasijevic, A, Sewall, L.M, Ward, A.B. | | 登録日 | 2020-12-31 | | 公開日 | 2021-08-04 | | 最終更新日 | 2021-09-08 | | 実験手法 | ELECTRON MICROSCOPY (3.66 Å) | | 主引用文献 | Polyclonal antibody responses to HIV Env immunogens resolved using cryoEM.
Nat Commun, 12, 2021
|
|
7M18
 
 | | HeLa-tubulin in complex with cryptophycin 1 | | 分子名称: | Cryptophycin 1, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Eren, E. | | 登録日 | 2021-03-12 | | 公開日 | 2021-09-08 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.38 Å) | | 主引用文献 | Conformational changes in tubulin upon binding cryptophycin-52 reveal its mechanism of action.
J.Biol.Chem., 297, 2021
|
|
7LXB
 
 | | HeLa-tubulin in complex with cryptophycin 52 | | 分子名称: | Cryptophycin 52, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Eren, E. | | 登録日 | 2021-03-03 | | 公開日 | 2021-09-08 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.26 Å) | | 主引用文献 | Conformational changes in tubulin upon binding cryptophycin-52 reveal its mechanism of action.
J.Biol.Chem., 297, 2021
|
|
7LOO
 
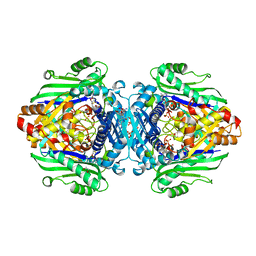 | | S-adenosyl methionine transferase cocrystallized with ATP | | 分子名称: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHOSPHATE ION, ... | | 著者 | Jackson, C.J, Tan, L.L, Laurino, P. | | 登録日 | 2021-02-10 | | 公開日 | 2021-09-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Substrate Dynamics Contribute to Enzymatic Specificity in Human and Bacterial Methionine Adenosyltransferases.
Jacs Au, 1, 2021
|
|
1NFV
 
 | | X-ray structure of Desulfovibrio desulfuricans bacterioferritin: the diiron centre in different catalytic states (as-isolated structure) | | 分子名称: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, 3-HYDROXYPYRUVIC ACID, FE (III) ION, ... | | 著者 | Macedo, S, Romao, C.V, Mitchell, E, Matias, P.M, Liu, M.Y, Xavier, A.V, LeGall, J, Teixeira, M, Lindley, P, Carrondo, M.A. | | 登録日 | 2002-12-16 | | 公開日 | 2003-04-01 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | The nature of the di-iron site in the bacterioferritin from
Desulfovibrio desulfuricans
NAT.STRUCT.BIOL., 10, 2003
|
|
7LS1
 
 | | 80S ribosome from mouse bound to eEF2 (Class II) | | 分子名称: | 28S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | 著者 | Loerch, S, Smith, P.R, Kunder, N, Stanowick, A.D, Lou, T.-F, Campbell, Z.T. | | 登録日 | 2021-02-17 | | 公開日 | 2021-11-03 | | 最終更新日 | 2021-12-08 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Functionally distinct roles for eEF2K in the control of ribosome availability and p-body abundance.
Nat Commun, 12, 2021
|
|
7LS2
 
 | | 80S ribosome from mouse bound to eEF2 (Class I) | | 分子名称: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | 著者 | Loerch, S, Smith, P.R, Kunder, N, Stanowick, A.D, Lou, T.-F, Campbell, Z.T. | | 登録日 | 2021-02-17 | | 公開日 | 2021-11-03 | | 最終更新日 | 2021-12-08 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Functionally distinct roles for eEF2K in the control of ribosome availability and p-body abundance.
Nat Commun, 12, 2021
|
|
7K00
 
 | | Structure of the Bacterial Ribosome at 2 Angstrom Resolution | | 分子名称: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | 著者 | Watson, Z.L, Ward, F.R, Meheust, R, Ad, O, Schepartz, A, Banfield, J.F, Cate, J.H.D. | | 登録日 | 2020-09-02 | | 公開日 | 2020-09-23 | | 最終更新日 | 2023-11-15 | | 実験手法 | ELECTRON MICROSCOPY (1.98 Å) | | 主引用文献 | Structure of the bacterial ribosome at 2 angstrom resolution.
Elife, 9, 2020
|
|
7K8T
 
 | | Structure of the SARS-CoV-2 S 6P trimer in complex with the human neutralizing antibody Fab fragment, C002 (State 2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C002 Fab Heavy Chain, ... | | 著者 | Malyutin, A.G, Barnes, C.O, Bjorkman, P.J. | | 登録日 | 2020-09-27 | | 公開日 | 2020-10-21 | | 最終更新日 | 2021-01-13 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7KE9
 
 | |
7K4N
 
 | |
1NMW
 
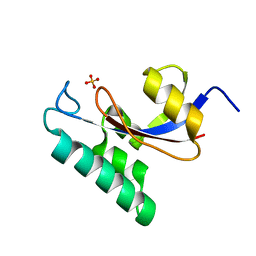 | | Solution structure of the PPIase domain of human Pin1 | | 分子名称: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, SULFATE ION | | 著者 | Bayer, E, Goettsch, S, Mueller, J.W, Griewel, B, Guiberman, E, Mayr, L, Bayer, P. | | 登録日 | 2003-01-12 | | 公開日 | 2003-07-15 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Analysis of the Mitotic Regulator hPin1 in Solution: INSIGHTS INTO DOMAIN ARCHITECTURE AND SUBSTRATE BINDING.
J.Biol.Chem., 278, 2003
|
|
7JT2
 
 | |
7JVC
 
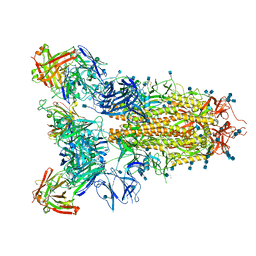 | | SARS-CoV-2 spike in complex with the S2A4 neutralizing antibody Fab fragment | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S2A4 Fab heavy chain, ... | | 著者 | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | 登録日 | 2020-08-20 | | 公開日 | 2020-10-14 | | 最終更新日 | 2021-06-23 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7JSV
 
 | | Cryo-EM structure of conjugative pili from carbapenem-resistant Klebsiella pneumoniae | | 分子名称: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, Pilin | | 著者 | Zheng, W, Pena, A, Frankel, G, Egelman, E.H. | | 登録日 | 2020-08-16 | | 公開日 | 2020-09-02 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cryoelectron-Microscopic Structure of the pKpQIL Conjugative Pili from Carbapenem-Resistant Klebsiella pneumoniae.
Structure, 28, 2020
|
|
7JWY
 
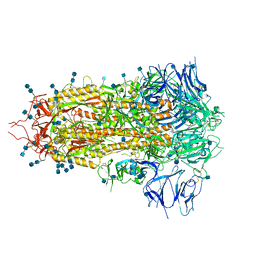 | | Structure of SARS-CoV-2 spike at pH 4.5 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhou, T, Tsybovsky, Y, Kwong, P.D. | | 登録日 | 2020-08-26 | | 公開日 | 2020-11-25 | | 最終更新日 | 2021-12-15 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7K3U
 
 | |
7K5I
 
 | |
7KDJ
 
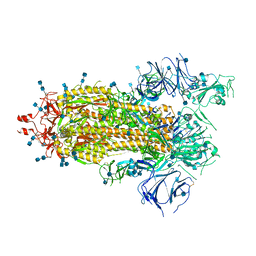 | |
7K4A
 
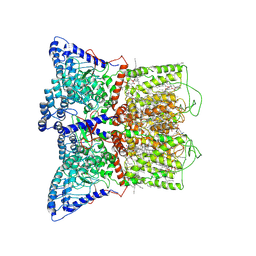 | | Cryo-EM structure of human TRPV6 in the open state | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, CHOLESTEROL HEMISUCCINATE, ... | | 著者 | Neuberger, A, Nadezhdin, K.D, Singh, A.K, Sobolevsky, A.I. | | 登録日 | 2020-09-15 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.26 Å) | | 主引用文献 | Inactivation-mimicking block of the epithelial calcium channel TRPV6.
Sci Adv, 6, 2020
|
|
7K5G
 
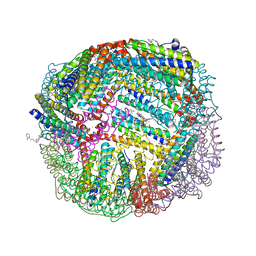 | | 1.95 A resolution structure of WT BfrB from Pseudomonas aeruginosa in complex with a protein-protein interaction inhibitor KM-5-28 | | 分子名称: | 4-{[3-(2-hydroxyphenyl)propyl]amino}-1H-isoindole-1,3(2H)-dione, Ferroxidase, POTASSIUM ION, ... | | 著者 | Lovell, S, Battaile, K.P, Soldano, A, Punchi-Hewage, A, Meraz, K, Annor-Gyamfi, J.K, Yao, H, Bunce, R.A, Rivera, M. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Small Molecule Inhibitors of the Bacterioferritin (BfrB)-Ferredoxin (Bfd) Complex Kill Biofilm-Embedded Pseudomonas aeruginosa Cells.
Acs Infect Dis., 7, 2021
|
|
7KE4
 
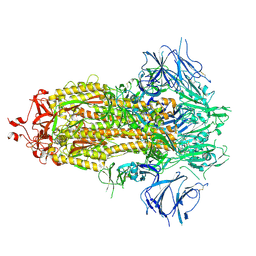 | |
