1QLR
 
 | | CRYSTAL STRUCTURE OF THE FAB FRAGMENT OF A HUMAN MONOCLONAL IgM COLD AGGLUTININ | | 分子名称: | IGM FAB REGION IV-J(H4)-C (KAU COLD AGGLUTININ), IGM KAPPA CHAIN V-III (KAU COLD AGGLUTININ), alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Carvalho, J.G, Cauerhff, A, Goldbaum, F, Leoni, J, Polikarpov, I. | | 登録日 | 1999-09-11 | | 公開日 | 2000-09-14 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.83 Å) | | 主引用文献 | Three-dimensional structure of the Fab from a human IgM cold agglutinin.
J Immunol., 165, 2000
|
|
2YI8
 
 | | Structure of the RNA polymerase VP1 from Infectious Pancreatic Necrosis Virus | | 分子名称: | CHLORIDE ION, POTASSIUM ION, RNA-DIRECTED RNA POLYMERASE | | 著者 | Graham, S.C, Sarin, L.P, Bahar, M.W, Myers, R.A, Stuart, D.I, Bamford, D.H, Grimes, J.M. | | 登録日 | 2011-05-11 | | 公開日 | 2011-07-20 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The N-Terminus of the RNA Polymerase from Infectious Pancreatic Necrosis Virus is the Determinant of Genome Attachment.
Plos Pathog., 7, 2011
|
|
3JWV
 
 | | Structure of rat neuronal nitric oxide synthase R349A mutant heme domain in complex with N1-{(3'S,4'R)-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-N2-(3'-fluorophenethyl)ethane-1,2-diamine | | 分子名称: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3S,4R)-4-[(6-amino-4-methylpyridin-2-yl)methyl]pyrrolidin-3-yl}-N'-[2-(3-fluorophenyl)ethyl]ethane-1,2-diamine, ... | | 著者 | Delker, S.L, Li, H, Poulos, T.L. | | 登録日 | 2009-09-18 | | 公開日 | 2010-05-05 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Unexpected binding modes of nitric oxide synthase inhibitors effective in the prevention of a cerebral palsy phenotype in an animal model.
J.Am.Chem.Soc., 132, 2010
|
|
3JX8
 
 | |
1QN1
 
 | | SOLUTION STRUCTURE OF DESULFOVIBRIO GIGAS FERRICYTOCHROME C3, NMR, 15 STRUCTURES | | 分子名称: | CYTOCHROME C3, HEME C | | 著者 | Brennan, L, Messias, A.C, Legall, J, Turner, D.L, Xavier, A.V. | | 登録日 | 1999-10-11 | | 公開日 | 2000-10-12 | | 最終更新日 | 2019-11-06 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural Basis for the Network of Functional Cooperativities in Cytochromes C3 from Desulfovibrio Gigas: Solution Structures of the Oxidised and Reduced States
J.Mol.Biol., 298, 2000
|
|
2YNA
 
 | |
5UE4
 
 | | proMMP-9desFnII complexed to JNJ0966 INHIBITOR | | 分子名称: | CALCIUM ION, Matrix metalloproteinase-9, SULFATE ION, ... | | 著者 | Alexander, R.S, Spurlino, J, Milligan, C. | | 登録日 | 2016-12-29 | | 公開日 | 2017-09-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Discovery of a highly selective chemical inhibitor of matrix metalloproteinase-9 (MMP-9) that allosterically inhibits zymogen activation.
J. Biol. Chem., 292, 2017
|
|
3JYR
 
 | | Crystal structures of the GacH receptor of Streptomyces glaucescens GLA.O in the unliganded form and in complex with acarbose and an acarbose homolog. Comparison with acarbose-loaded maltose binding protein of Salmonella typhimurium. | | 分子名称: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Maltose-binding periplasmic protein | | 著者 | Vahedi-Faridi, A, Licht, A, Bulut, H, Schneider, E. | | 登録日 | 2009-09-22 | | 公開日 | 2010-02-16 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal Structures of the Solute Receptor GacH of Streptomyces glaucescens in Complex with Acarbose and an Acarbose Homolog: Comparison with the Acarbose-Loaded Maltose-Binding Protein of Salmonella typhimurium.
J.Mol.Biol., 397, 2010
|
|
5FYU
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with 3D fragment 3-Amino-4-methyl-1,3-dihydro-2H-indol-2-one (N10042a) | | 分子名称: | (3R)-3-azanyl-4-methyl-1,3-dihydroindol-2-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | 著者 | Nowak, R, Krojer, T, Johansson, C, Pearce, N, Gileadi, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, von Delft, F, Brennan, P.E, Oppermann, U. | | 登録日 | 2016-03-09 | | 公開日 | 2016-03-23 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with N10042A
To be Published
|
|
2YG2
 
 | | Structure of apolioprotein M in complex with Sphingosine 1-Phosphate | | 分子名称: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, APOLIPOPROTEIN M, CITRATE ANION | | 著者 | Christoffersen, C, Obinata, H, Gowda, S, Galvani, S, Ahnstrom, J, Sevvana, M, Egerer-Sieber, C, Muller, Y.A, Hla, T, Nielsen, L, Dahlback, B. | | 登録日 | 2011-04-11 | | 公開日 | 2011-06-01 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Endothelium-Protective Sphingosine-1-Phosphate Provided by Hdl-Associated Apolipoprotein M.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5FZM
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with 3D fragment 5-(2-fluorophenyl)-1,3-oxazole-4-carboxylic acid (N09989b) (ligand modelled based on PANDDA event map, SGC - Diamond I04-1 fragment screening) | | 分子名称: | 1,2-ETHANEDIOL, 5-(2-fluorophenyl)-1,3-oxazole-4-carboxylic acid, CHLORIDE ION, ... | | 著者 | Nowak, R, Krojer, T, Johansson, C, Kupinska, K, Szykowska, A, Pearce, N, Talon, R, Collins, P, Gileadi, C, Strain-Damerell, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, von Delft, F, Brennan, P.E, Oppermann, U. | | 登録日 | 2016-03-14 | | 公開日 | 2017-03-29 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.49 Å) | | 主引用文献 | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with 3D Fragment 5-(2-Fluorophenyl)-1,3-Oxazole-4-Carboxylic Acid (N09989B) (Ligand Modelled Based on Pandda Event Map, Sgc - Diamond I04-1 Fragment Screening)
To be Published
|
|
2YI9
 
 | | Structure of the RNA polymerase VP1 from Infectious Pancreatic Necrosis Virus in complex with magnesium | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, POTASSIUM ION, ... | | 著者 | Graham, S.C, Sarin, L.P, Bahar, M.W, Myers, R.A, Stuart, D.I, Bamford, D.H, Grimes, J.M. | | 登録日 | 2011-05-11 | | 公開日 | 2011-07-20 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | The N-Terminus of the RNA Polymerase from Infectious Pancreatic Necrosis Virus is the Determinant of Genome Attachment.
Plos Pathog., 7, 2011
|
|
2N4O
 
 | |
3JZU
 
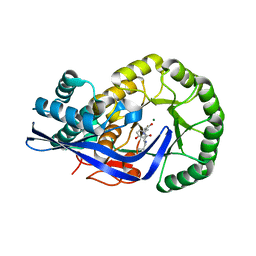 | | Crystal structure of Dipeptide Epimerase from Enterococcus faecalis V583 complexed with Mg and dipeptide L-Leu-L-Tyr | | 分子名称: | Dipeptide Epimerase, LEUCINE, MAGNESIUM ION, ... | | 著者 | Fedorov, A.A, Fedorov, E.V, Imker, H.J, Sakai, A, Gerlt, J.A, Almo, S.C. | | 登録日 | 2009-09-24 | | 公開日 | 2010-08-18 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2N7A
 
 | | Solution structure of the human Siglec-8 lectin domain | | 分子名称: | Sialic acid-binding Ig-like lectin 8 | | 著者 | Proepster, J.M, Yang, F, Rabbani, S, Ernst, B, Allain, F.H.-T, Schubert, M. | | 登録日 | 2015-09-07 | | 公開日 | 2016-07-06 | | 最終更新日 | 2023-06-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis for sulfation-dependent self-glycan recognition by the human immune-inhibitory receptor Siglec-8.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
2YBJ
 
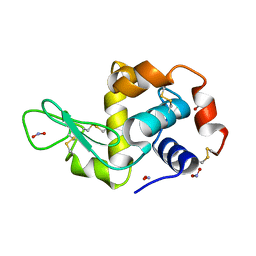 | |
3K0X
 
 | | Crystal structure of telomere capping protein Ten1 from Saccharomyces pombe | | 分子名称: | IODIDE ION, Protein Ten1 | | 著者 | Gelinas, A.D, Reyes, F.E, Batey, R.T, Wuttke, D.S. | | 登録日 | 2009-09-25 | | 公開日 | 2009-10-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Telomere capping proteins are structurally related to RPA with an additional telomere-specific domain.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5FZ8
 
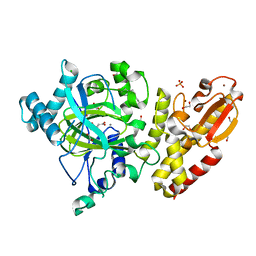 | | Crystal structure of the catalytic domain of human JARID1B in complex with malate | | 分子名称: | (2S)-2-hydroxybutanedioic acid, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Nowak, R, Krojer, T, Johansson, C, Gileadi, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, von Delft, F, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | 登録日 | 2016-03-11 | | 公開日 | 2017-03-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with Malate
To be Published
|
|
2YBQ
 
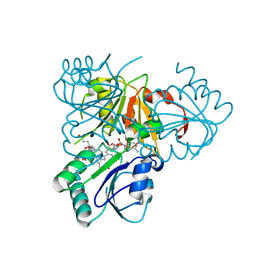 | | The x-ray structure of the SAM-dependent uroporphyrinogen III methyltransferase NirE from Pseudomonas aeruginosa in complex with SAH and uroporphyrinogen III | | 分子名称: | METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE, UROPORPHYRINOGEN III | | 著者 | Storbeck, S, Saha, S, Krausze, J, Klink, B.U, Heinz, D.W, Layer, G. | | 登録日 | 2011-03-09 | | 公開日 | 2011-06-01 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of the Heme D1 Biosynthesis Enzyme Nire in Complex with its Substrate Reveals New Insights Into the Catalytic Mechanism of S-Adenosyl-L-Methionine-Dependent Uroporphyrinogen III Methyltransferases.
J.Biol.Chem., 286, 2011
|
|
2NC0
 
 | |
3K11
 
 | |
2YCS
 
 | | Crystal structure of checkpoint kinase 2 in complex with PV788 | | 分子名称: | N-{4-[(1E)-N-CARBAMIMIDOYLETHANEHYDRAZONOYL]PHENYL}-1H-INDOLE-3-CARBOXAMIDE, NITRATE ION, SERINE/THREONINE-PROTEIN KINASE CHK2 | | 著者 | Lountos, G.T, Jobson, A.G, Tropea, J.E, Self, C.R, Pommier, Y, Shoemaker, R.H, Zhang, G, Waugh, D.S. | | 登録日 | 2011-03-16 | | 公開日 | 2011-11-16 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural Characterization of Inhibitor Complexes with Checkpoint Kinase 2 (Chk2), a Drug Target for Cancer Therapy.
J.Struct.Biol., 176, 2011
|
|
2NC1
 
 | |
5TPG
 
 | | Optimization of spirocyclic proline tryptophanhydroxylase-1 inhibitors | | 分子名称: | (3S)-8-(2-amino-6-{(1R)-1-[5-chloro-3'-(methylsulfonyl)[1,1'-biphenyl]-2-yl]-2,2,2-trifluoroethoxy}pyrimidin-4-yl)-2,8-diazaspiro[4.5]decane-3-carboxylic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETONITRILE, ... | | 著者 | Stein, A.J, Goldberg, D.R, De Lombaert, S, Holt, M.C. | | 登録日 | 2016-10-20 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Optimization of spirocyclic proline tryptophan hydroxylase-1 inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5TRF
 
 | | MDM2 in complex with SAR405838 | | 分子名称: | (2'S,3R,4'S,5'R)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-N-(trans-4-hydroxycyclohexyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide, E3 ubiquitin-protein ligase Mdm2, GLYCEROL, ... | | 著者 | Meagher, J.L, Stuckey, J.A. | | 登録日 | 2016-10-26 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | SAR405838: an optimized inhibitor of MDM2-p53 interaction that induces complete and durable tumor regression.
Cancer Res., 74, 2014
|
|
