6FX1
 
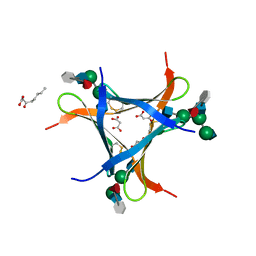 | | Crystal structure of Pholiota squarrosa lectin in complex with an octasaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]1-azido-beta-N-acetyl-D-glucosamine, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]1-azido-beta-N-acetyl-D-glucosamine, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]1-azido-beta-N-acetyl-D-glucosamine, ... | | Authors: | Cabanettes, A, Varrot, A. | | Deposit date: | 2018-03-08 | | Release date: | 2018-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Recognition of Complex Core-Fucosylated N-Glycans by a Mini Lectin.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
7LQX
 
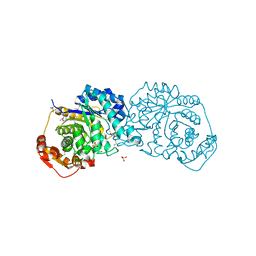 | |
6K8K
 
 | | Crystal structure of Arabidopsis thaliana BIC2-CRY2 complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Cryptochrome-2, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Wang, X, Ma, L, Guan, Z, Yin, P. | | Deposit date: | 2019-06-12 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into BIC-mediated inactivation of Arabidopsis cryptochrome 2.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6KAY
 
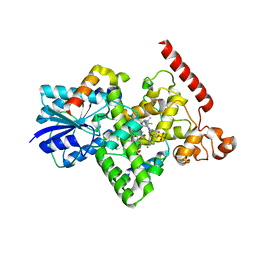
 | | X-ray structure of human PPARalpha ligand binding domain-GW7647 co-crystals obtained by soaking | | Descriptor: | 2-[(4-{2-[(4-cyclohexylbutyl)(cyclohexylcarbamoyl)amino]ethyl}phenyl)sulfanyl]-2-methylpropanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Suda, K, Saito, K, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.735 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
8PKB
 
 | |
7LR1
 
 | |
6DEX
 
 | | Structure of Eremothecium gossypii Shu1:Shu2 complex | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, MAGNESIUM ION, Suppressor of hydroxyurea sensitivity protein 1, ... | | Authors: | Singh, N, Corbett, K.D. | | Deposit date: | 2018-05-13 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Eremothecium gossypii Shu1:Shu2 complex
To Be Published
|
|
5WED
 
 | |
6KB3
 
 | | X-ray structure of human PPARalpha ligand binding domain-GW7647 co-crystals obtained by delipidation and cross-seeding | | Descriptor: | 2-[(4-{2-[(4-cyclohexylbutyl)(cyclohexylcarbamoyl)amino]ethyl}phenyl)sulfanyl]-2-methylpropanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor alpha | | Authors: | Kamata, S, Saito, K, Honda, A, Ishikawa, R, Oyama, T, Ishii, I. | | Deposit date: | 2019-06-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | PPAR alpha Ligand-Binding Domain Structures with Endogenous Fatty Acids and Fibrates.
Iscience, 23, 2020
|
|
8PM5
 
 | |
5FTT
 
 | | Octameric complex of Latrophilin 3 (Lec, Olf) , Unc5D (Ig, Ig2, TSP1) and FLRT2 (LRR) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADHESION G PROTEIN-COUPLED RECEPTOR L3, CALCIUM ION, ... | | Authors: | Jackson, V.A, Mehmood, S, Chavent, M, Roversi, P, Carrasquero, M, del Toro, D, Seyit-Bremer, G, Ranaivoson, F.M, Comoletti, D, Sansom, M.S.P, Robinson, C.V, Klein, R, Seiradake, E. | | Deposit date: | 2016-01-15 | | Release date: | 2016-05-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Super-Complexes of Adhesion Gpcrs and Neural Guidance Receptors
Nat.Commun., 7, 2016
|
|
7RLK
 
 | | Wallaby TTR | | Descriptor: | Transthyretin, ZINC ION | | Authors: | D-Souza, D.G, Richardson, S.J. | | Deposit date: | 2021-07-25 | | Release date: | 2022-08-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural and amyloidogenic comparisons of human and wallaby transthyretins: implications for amyloidosis?
To Be Published
|
|
7LSG
 
 | |
7ROF
 
 | |
6FQV
 
 | | 2.60A BINARY COMPLEX OF S.AUREUS GYRASE with UNCLEAVED DNA | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*GP*TP*AP*CP*GP*GP*CP*CP*GP*TP*AP*CP*GP*CP*TP*T)-3'), DNA gyrase subunit A, DNA gyrase subunit B,DNA gyrase subunit B, ... | | Authors: | Bax, B.D, Germe, T, Basque, E, Maxwell, A. | | Deposit date: | 2018-02-14 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A new class of antibacterials, the imidazopyrazinones, reveal structural transitions involved in DNA gyrase poisoning and mechanisms of resistance.
Nucleic Acids Res., 46, 2018
|
|
7RMX
 
 | | Structure of De Novo designed tunable symmetric protein pockets | | Descriptor: | Tunable symmetric protein, D_3_212 | | Authors: | Bera, A.K, Hicks, D.R, Kang, A, Sankaran, B, Baker, D. | | Deposit date: | 2021-07-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | De novo design of protein homodimers containing tunable symmetric protein pockets.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6DFI
 
 | |
8TNN
 
 | | Crystal structure of Epstein-Barr virus gH/gL/gp42 in complex with gp42 antibody A10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, A10 heavy chain, A10 light chain, ... | | Authors: | Bu, W, Kumar, A, Board, N, Kim, J, Dowdell, K, Zhang, S, Lei, Y, Hostal, A, Krogmann, T, Wang, Y, Pittaluga, S, Marcotrigiano, J, Cohen, J.I. | | Deposit date: | 2023-08-02 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Epstein-Barr virus gp42 antibodies reveal sites of vulnerability for receptor binding and fusion to B cells.
Immunity, 57, 2024
|
|
7LR8
 
 | |
6G09
 
 | | Crystal Structure of a GH8 xylobiose complex from Teredinibacter turnerae | | Descriptor: | 1,2-ETHANEDIOL, Glycoside hydrolase family 8 domain protein, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Fowler, C.A, Davies, G.J, Walton, P.H. | | Deposit date: | 2018-03-16 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and function of a glycoside hydrolase family 8 endoxylanase from Teredinibacter turnerae.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
7RNQ
 
 | |
5FF5
 
 | | Crystal Structure of SeMet PaaA | | Descriptor: | GLYCEROL, NICKEL (II) ION, PaaA, ... | | Authors: | Biernat, K.B, Redinbo, M.R. | | Deposit date: | 2015-12-17 | | Release date: | 2016-04-27 | | Last modified: | 2016-05-18 | | Method: | X-RAY DIFFRACTION (2.933 Å) | | Cite: | Post-translational Claisen Condensation and Decarboxylation en Route to the Bicyclic Core of Pantocin A.
J.Am.Chem.Soc., 138, 2016
|
|
5FW2
 
 | | Crystal structure of SpCas9 variant EQR bound to sgRNA and TGAG PAM target DNA | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MAGNESIUM ION, NON-TARGET DNA STRAND, ... | | Authors: | Anders, C, Bargsten, K, Jinek, M. | | Deposit date: | 2016-02-11 | | Release date: | 2016-06-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.676 Å) | | Cite: | Structural Plasticity of Pam Recognition by Engineered Variants of the RNA-Guided Endonuclease Cas9.
Mol.Cell, 61, 2016
|
|
8TCW
 
 | |
6P5U
 
 | | Structure of an enoyl-CoA hydratase/aldolase isolated from a lignin-degrading consortium | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COENZYME A, Enoyl-CoA hydratase | | Authors: | Liberato, M.V, Squina, F.M. | | Deposit date: | 2019-05-31 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of a prokaryotic feruloyl-CoA hydratase-lyase from a lignin-degrading consortium with high oligomerization stability under extreme pHs.
Biochim Biophys Acta Proteins Proteom, 1868, 2020
|
|
