6E8D
 
 | |
3B99
 
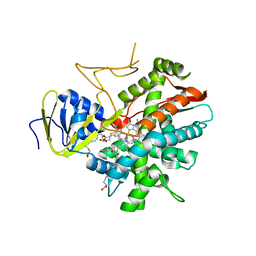 | | Crystal structure of zebrafish prostacyclin synthase (cytochrome P450 8A1) in complex with substrate analog U51605 | | Descriptor: | (5Z)-7-{(1R,4S,5R,6R)-6-[(1E)-oct-1-en-1-yl]-2,3-diazabicyclo[2.2.1]hept-2-en-5-yl}hept-5-enoic acid, PROTOPORPHYRIN IX CONTAINING FE, Prostaglandin I2 synthase | | Authors: | Li, Y.-C, Chiang, C.-W, Yeh, H.-C, Hsu, P.-Y, Whitby, F.G, Wang, L.-H, Chan, N.-L. | | Deposit date: | 2007-11-03 | | Release date: | 2007-11-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Prostacyclin Synthase and Its Complexes with Substrate Analog and Inhibitor Reveal a Ligand-specific Heme Conformation Change
J.Biol.Chem., 283, 2008
|
|
3X22
 
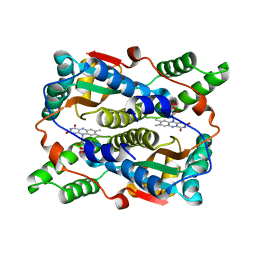 | | Crystal structure of Escherichia coli nitroreductase NfsB mutant N71S/F123A/F124W | | Descriptor: | FLAVIN MONONUCLEOTIDE, Oxygen-insensitive NAD(P)H nitroreductase | | Authors: | Bai, J, Yang, J, Zhou, Y, Yang, Q. | | Deposit date: | 2014-12-06 | | Release date: | 2015-11-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis of Escherichia coli nitroreductase NfsB triple mutants engineered for improved activity and regioselectivity toward the prodrug CB1954
PROCESS BIOCHEM, 50, 2015
|
|
1LF4
 
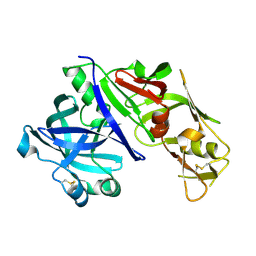 | | STRUCTURE OF PLASMEPSIN II | | Descriptor: | Plasmepsin 2 | | Authors: | Asojo, O.A, Gulnik, S.V, Afonina, E, Yu, B, Ellman, J.A, Haque, T.S, Silva, A.M. | | Deposit date: | 2002-04-10 | | Release date: | 2002-10-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel uncomplexed and complexed structures of plasmepsin II, an aspartic protease from Plasmodium falciparum.
J.Mol.Biol., 327, 2003
|
|
3BDG
 
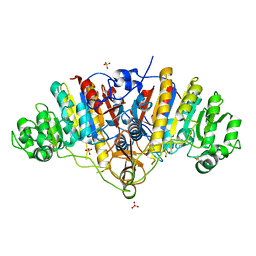 | |
3MOG
 
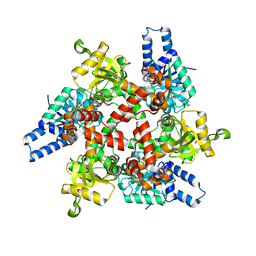 | | Crystal structure of 3-hydroxybutyryl-CoA dehydrogenase from Escherichia coli K12 substr. MG1655 | | Descriptor: | CHLORIDE ION, GLYCEROL, Probable 3-hydroxybutyryl-CoA dehydrogenase | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Gilmore, M, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-04-22 | | Release date: | 2010-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of 3-Hydroxybutyryl-Coa Dehydrogenase from Escherichia Coli K12
To be Published
|
|
3X0Y
 
 | | Crystal structure of FMN-bound DszC from Rhodococcus erythropolis D-1 | | Descriptor: | DszC, FLAVIN MONONUCLEOTIDE | | Authors: | Guan, L.J, Lee, W.C, Wang, S.P, Ohtsuka, J, Tanokura, M. | | Deposit date: | 2014-10-23 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of apo-DszC and FMN-bound DszC from Rhodococcus erythropolis D-1.
Febs J., 282, 2015
|
|
3BDF
 
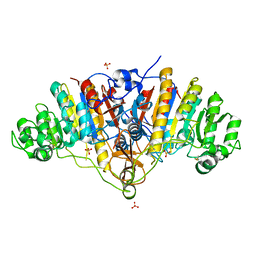 | |
6ECM
 
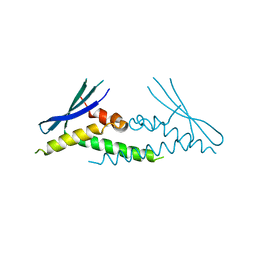 | |
1GW1
 
 | | Substrate distortion by beta-mannanase from Pseudomonas cellulosa | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DINITROPHENYLENE, MANNAN ENDO-1,4-BETA-MANNOSIDASE, ... | | Authors: | Ducros, V, Zechel, D.L, Gilbert, H.J, Szabo, L, Withers, S.G, Davies, G.J. | | Deposit date: | 2002-03-01 | | Release date: | 2002-09-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Substrate Distortion by a Beta-Mannanase: Snapshots of the Michaelis and Covalent-Intermediate Complexes Suggest a B2,5 Conformation for the Transition State
Angew.Chem.Int.Ed.Engl., 41, 2002
|
|
1H13
 
 | | Structure of a cold-adapted family 8 xylanase | | Descriptor: | ENDO-1,4-BETA-XYLANASE | | Authors: | Van Petegem, F, Collins, T, Meuwis, M.A, Feller, G, Gerday, C, Van Beeumen, J. | | Deposit date: | 2002-07-02 | | Release date: | 2003-03-13 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Structure of a Cold-Adapted Family 8 Xylanase at 1.3 A Resolution: Structural Adaptations to Cold and Investigation of the Active Site
J.Biol.Chem., 278, 2003
|
|
3MAS
 
 | |
3BF7
 
 | |
4JTN
 
 | |
3WZW
 
 | |
3BG6
 
 | | Pyranose 2-oxidase from Trametes multicolor, E542K mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Pyranose oxidase | | Authors: | Tan, T.C, Divne, C. | | Deposit date: | 2007-11-26 | | Release date: | 2008-12-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Improving thermostability and catalytic activity of pyranose 2-oxidase from Trametes multicolor by rational and semi-rational design
Febs J., 276, 2009
|
|
3WXU
 
 | |
3WY1
 
 | | Crystal structure of alpha-glucosidase | | Descriptor: | (3R,5R,7R)-octane-1,3,5,7-tetracarboxylic acid, Alpha-glucosidase, GLYCEROL, ... | | Authors: | Shen, X, Gai, Z, Kato, K, Yao, M. | | Deposit date: | 2014-08-18 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural analysis of the alpha-glucosidase HaG provides new insights into substrate specificity and catalytic mechanism
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
4JWU
 
 | | Crystal structure of Cytochrome P450cam-putidaredoxin complex | | Descriptor: | 1,1'-hexane-1,6-diyldipyrrolidine-2,5-dione, CALCIUM ION, Camphor 5-monooxygenase, ... | | Authors: | Tripathi, S.M, Li, H, Poulos, T.L. | | Deposit date: | 2013-03-27 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for effector control and redox partner recognition in cytochrome P450.
Science, 340, 2013
|
|
1CC6
 
 | | PHE161 AND ARG166 VARIANTS OF P-HYDROXYBENZOATE HYDROXYLASE. IMPLICATIONS FOR NADPH RECOGNITION AND STRUCTURAL STABILITY. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID, PROTEIN (P-HYDROXYBENZOATE HYDROXYLASE) | | Authors: | Eppink, M.H.M, Bunthof, C, Schreuder, H.A, Van Berkel, W.J.H. | | Deposit date: | 1999-03-04 | | Release date: | 1999-03-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Phe161 and Arg166 variants of p-hydroxybenzoate hydroxylase. Implications for NADPH recognition and structural stability.
Febs Lett., 443, 1999
|
|
3MDL
 
 | | X-ray crystal structure of 1-arachidonoyl glycerol bound to the cyclooxygenase channel of cyclooxygenase-2 | | Descriptor: | (2S)-2,3-dihydroxypropyl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vecchio, A.J, Malkowski, M.G. | | Deposit date: | 2010-03-30 | | Release date: | 2011-04-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structural basis of endocannabinoid oxygenation by cyclooxygenase-2.
J.Biol.Chem., 286, 2011
|
|
3ZFV
 
 | | Crystal structure of an archaeal CRISPR-associated Cas6 nuclease | | Descriptor: | CRISPR-ASSOCIATED ENDORIBONUCLEASE CAS6 1, GLYCEROL | | Authors: | Reeks, J, Liu, H, White, M.F, Naismith, J.H. | | Deposit date: | 2012-12-12 | | Release date: | 2013-04-03 | | Last modified: | 2013-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a Dimeric Crenarchaeal Cas6 Enzyme with an Atypical Active Site for Crispr RNA Processing
Biochem.J., 452, 2013
|
|
8B3Q
 
 | |
3B1J
 
 | | Crystal structure of Glyceraldehyde-3-Phosphate Dehydrogenase complexed with CP12 in the presence of copper from Synechococcus elongatus | | Descriptor: | COPPER (II) ION, CP12, Glyceraldehyde 3-phosphate dehydrogenase (NADP+), ... | | Authors: | Matsumura, H, Kai, A, Inoue, T. | | Deposit date: | 2011-07-04 | | Release date: | 2012-01-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure Basis for the Regulation of Glyceraldehyde-3-Phosphate Dehydrogenase Activity via the Intrinsically Disordered Protein CP12.
Structure, 19, 2011
|
|
1L4Z
 
 | | X-RAY CRYSTAL STRUCTURE OF THE COMPLEX OF MICROPLASMINOGEN WITH ALPHA DOMAIN OF STREPTOKINASE IN THE PRESENCE CADMIUM IONS | | Descriptor: | CADMIUM ION, Plasminogen, Streptokinase | | Authors: | Wakeham, N, Terzyan, S, Zhai, P, Loy, J.A, Tang, J, Zhang, X.C. | | Deposit date: | 2002-03-06 | | Release date: | 2002-12-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Effects of deletion of streptokinase residues 48-59 on plasminogen activation.
PROTEIN ENG., 15, 2002
|
|
