8B9S
 
 | |
7R3D
 
 | | CRYSTAL STRUCTURE OF E.coli ALCOHOL DEHYDROGENASE - FucO MUTANT N151G, L259V COMPLEXED WITH FE, NADH, AND GLYCEROL (Absence of Nicotinamide ring) | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, FE (III) ION, Lactaldehyde reductase | | Authors: | Sridhar, S, Kiema, T.R, Wierenga, R, Widersten, M. | | Deposit date: | 2022-02-07 | | Release date: | 2022-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of lactaldehyde reductase, FucO, link enzyme activity to hydrogen bond networks and conformational dynamics.
Febs J., 290, 2023
|
|
7M03
 
 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 18c | | Descriptor: | (1R,2S)-2-((S)-2-((((3-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-((((3-fluorobenzyl)oxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Kashipathy, M.M, Lovell, S, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
8B9T
 
 | |
6R35
 
 | | Structure of the LecB lectin from Pseudomonas aeruginosa strain PAO1 in complex with lewis x tetrasaccharide | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Fucose-binding lectin PA-IIL, ... | | Authors: | Lepsik, M, Sommer, R, Kuhaudomlarp, S, Lelimousin, M, Varrot, A, Titz, A, Imberty, A. | | Deposit date: | 2019-03-19 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Induction of rare conformation of oligosaccharide by binding to calcium-dependent bacterial lectin: X-ray crystallography and modelling study.
Eur.J.Med.Chem., 177, 2019
|
|
8BV9
 
 | | Acylphosphatase from E. coli | | Descriptor: | 1,2-ETHANEDIOL, Acylphosphatase, GLYCEROL, ... | | Authors: | Gavira, J.A, Martinez-Rodriguez, S. | | Deposit date: | 2023-01-18 | | Release date: | 2023-10-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | First 3-D structural evidence of a native-like intertwined dimer in the acylphosphatase family.
Biochem.Biophys.Res.Commun., 682, 2023
|
|
8PWX
 
 | |
6DUJ
 
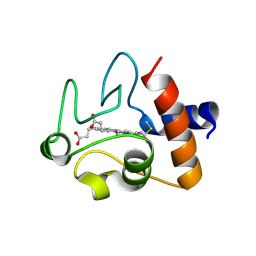 | | Crystal structure of A51V variant of Human Cytochrome c | | Descriptor: | Cytochrome c, HEME C | | Authors: | Lei, H, Bowler, B.E. | | Deposit date: | 2018-06-20 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.82202721 Å) | | Cite: | Naturally Occurring A51V Variant of Human CytochromecDestabilizes the Native State and Enhances Peroxidase Activity.
J.Phys.Chem.B, 123, 2019
|
|
6GQ0
 
 | | Crystal structure of GanP, a glucose-galactose binding protein from Geobacillus stearothermophilus | | Descriptor: | Putative sugar binding protein | | Authors: | Sherf, D, Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | Deposit date: | 2018-06-07 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The crystal structure of GanP, a glucose-galactose binding protein from Gebacillus Stearothermophilus
To Be Published
|
|
8B82
 
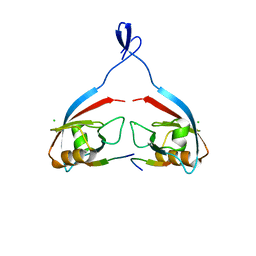 | |
6R3X
 
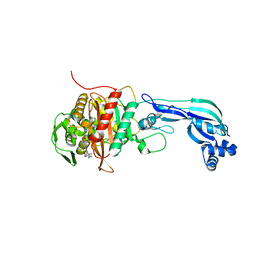 | |
6GMM
 
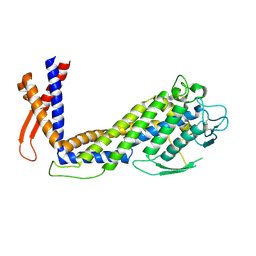 | | Crystal structure of Helicobacter pylori adhesin LabA | | Descriptor: | adhesin LabA | | Authors: | Paraskevopoulou, V, Overman, R.C, Stolnik, S, Winkler, S, Gellert, P, Falcone, F.H, Schimpl, M. | | Deposit date: | 2018-05-27 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural and binding characterization of the LacdiNAc-specific adhesin (LabA; HopD) exodomain from Helicobacter pylori
Current Research in Structural Biology, 2021
|
|
8B8O
 
 | |
6R42
 
 | |
8BL3
 
 | |
6GQD
 
 | | Structure of human galactose-1-phosphate uridylyltransferase (GALT), with crystallization epitope mutations A21Y:A22T:T23P:R25L | | Descriptor: | 1,2-ETHANEDIOL, 5,6-DIHYDROURIDINE-5'-MONOPHOSPHATE, Galactose-1-phosphate uridylyltransferase, ... | | Authors: | Fairhead, M, Strain-Damerell, C, Kopec, J, Bezerra, G.A, Zhang, M, Burgess-Brown, N, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-07 | | Release date: | 2018-07-18 | | Method: | X-RAY DIFFRACTION (1.523 Å) | | Cite: | Structure of human galactose-1-phosphate uridylyltransferase (GALT), with crystallization epitope mutations A21Y:A22T:T23P:R25L
To Be Published
|
|
6DJQ
 
 | | Vps1 GTPase-BSE fusion complexed with GDP.AlF4- | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Varlakhanova, N.V, Brady, T.M, Tornabene, B.A, Hosford, C.J, Chappie, J.S, Ford, M.G.J. | | Deposit date: | 2018-05-25 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the fungal dynamin-related protein Vps1 reveal a unique, open helical architecture.
J. Cell Biol., 217, 2018
|
|
6R4A
 
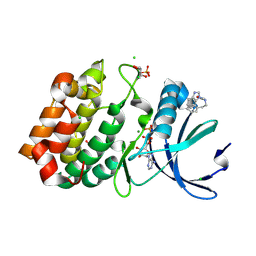 | | Aurora-A in complex with shape-diverse fragment 55 | | Descriptor: | 2-(benzimidazol-1-yl)-~{N}-(2-phenylethyl)ethanamide, ADENOSINE-5'-DIPHOSPHATE, Aurora kinase A, ... | | Authors: | Bayliss, R, McIntyre, P.J. | | Deposit date: | 2019-03-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.937 Å) | | Cite: | Construction of a Shape-Diverse Fragment Set: Design, Synthesis and Screen against Aurora-A Kinase.
Chemistry, 25, 2019
|
|
5WRP
 
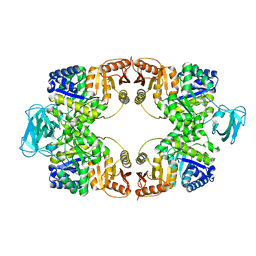 | | T-state crystal structure of pyruvate kinase from Mycobacterium tuberculosis | | Descriptor: | PHOSPHATE ION, Pyruvate kinase | | Authors: | Zhong, W, Cai, Q, El Sahili, A, Lescar, J, Dedon, P.C. | | Deposit date: | 2016-12-02 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Allosteric pyruvate kinase-based "logic gate" synergistically senses energy and sugar levels in Mycobacterium tuberculosis.
Nat Commun, 8, 2017
|
|
8BJS
 
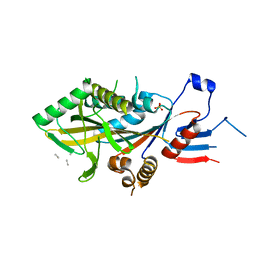 | | Apo KIF20A[55-510] crystal structure | | Descriptor: | Kinesin-like protein KIF20A, SULFATE ION, UNK-UNK-UNK-UNK | | Authors: | Ranaivoson, F.M, Kikuti, C, Crozet, V, Sirkia, M.E, Houdusse, A. | | Deposit date: | 2022-11-06 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Nucleotide-free structures of KIF20A illuminate atypical mechanochemistry in this kinesin-6.
Open Biology, 13, 2023
|
|
7RML
 
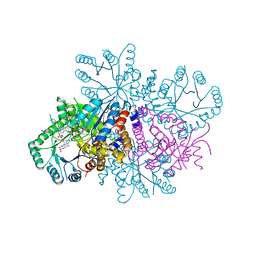 | | Neisseria meningitidis Methylenetetrahydrofolate reductase in complex with FAD | | Descriptor: | 5,10-methylenetetrahydrofolate reductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Pederick, J.L, Wegener, K.L, Salaemae, W, Bruning, J.B. | | Deposit date: | 2021-07-27 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Biochemical and structural characterization of meningococcal methylenetetrahydrofolate reductase.
Protein Sci., 32, 2023
|
|
7LZY
 
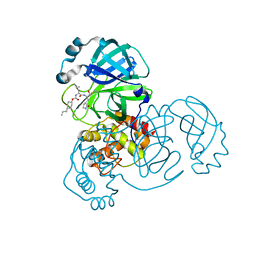 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 3c | | Descriptor: | (1R,2S)-1-hydroxy-2-((S)-4-methyl-2-(((((1s,4S)-4-propylcyclohexyl)oxy)carbonyl)amino)pentanamido)-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-1-hydroxy-2-((S)-4-methyl-2-(((((1s,4S)-4-propylcyclohexyl)oxy)carbonyl)amino)pentanamido)-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
6ISW
 
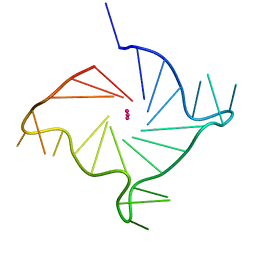 | | Structure of human telomeric DNA with 5-selenophene-modified deoxyuridine at residue 12 | | Descriptor: | DNA (22-MER), POTASSIUM ION | | Authors: | Saikrishnan, K, Nuthanakanti, A, Srivatsan, S.G, Ahmad, I. | | Deposit date: | 2018-11-19 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Probing G-quadruplex topologies and recognition concurrently in real time and 3D using a dual-app nucleoside probe.
Nucleic Acids Res., 47, 2019
|
|
7M01
 
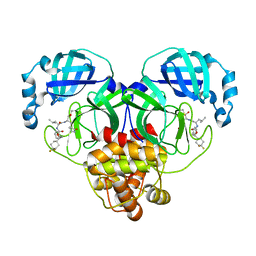 | | Structure of SARS-CoV-2 3CL protease in complex with inhibitor 14c | | Descriptor: | (1R,2S)-2-((S)-2-((((S)-1-(4,4-difluorocyclohexyl)-2-phenylethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-((((S)-1-(4,4-difluorocyclohexyl)-2-phenylethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Kim, Y, Perera, K.D, Jesri, A.R.M, Nguyen, H.N, Baird, M.A, Miller, M.J, Groutas, W.C, Chang, K.O. | | Deposit date: | 2021-03-10 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies.
J.Med.Chem., 64, 2021
|
|
8BL6
 
 | |
