6RAS
 
 | | Pmar-Lig_Pre. | | Descriptor: | ADENOSINE MONOPHOSPHATE, ATP-dependent DNA ligase, DNA, ... | | Authors: | Leiros, H.K.S, Williamson, A. | | Deposit date: | 2019-04-07 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural intermediates of a DNA-ligase complex illuminate the role of the catalytic metal ion and mechanism of phosphodiester bond formation.
Nucleic Acids Res., 47, 2019
|
|
6FPG
 
 | | Structure of the Ustilago maydis chorismate mutase 1 in complex with a Zea mays kiwellin | | Descriptor: | CITRIC ACID, Chromosome 16, whole genome shotgun sequence, ... | | Authors: | Altegoer, F, Steinchen, W, Bange, G. | | Deposit date: | 2018-02-09 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A kiwellin disarms the metabolic activity of a secreted fungal virulence factor.
Nature, 565, 2019
|
|
8QDY
 
 | |
5HW5
 
 | |
7RGY
 
 | |
6DBN
 
 | | Jak1 with compound 23 | | Descriptor: | Tyrosine-protein kinase JAK1, [(1S)-2,2-difluorocyclopropyl][(1R,5S)-3-{2-[(1-methyl-1H-pyrazol-4-yl)amino]pyrimidin-4-yl}-3,8-diazabicyclo[3.2.1]octan-8-yl]methanone | | Authors: | Vajdos, F.F. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-29 | | Last modified: | 2018-11-07 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Dual Inhibition of TYK2 and JAK1 for the Treatment of Autoimmune Diseases: Discovery of (( S)-2,2-Difluorocyclopropyl)((1 R,5 S)-3-(2-((1-methyl-1 H-pyrazol-4-yl)amino)pyrimidin-4-yl)-3,8-diazabicyclo[3.2.1]octan-8-yl)methanone (PF-06700841).
J. Med. Chem., 61, 2018
|
|
7MWC
 
 | | ERAP1 binds peptide C-terminus of a LPF sequence (AAAAFKARKF) | | Descriptor: | Endoplasmic reticulum aminopeptidase 1,LPF sequence | | Authors: | Guo, H.C, Sui, L. | | Deposit date: | 2021-05-16 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | ERAP1 binds peptide C-termini of different sequences and/or lengths by a common recognition mechanism.
Immunobiology, 226, 2021
|
|
5TVX
 
 | |
6IH1
 
 | | Crystal structure of a standalone versatile EAL protein from Vibrio cholerae O395 - c-di-GMP bound form | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, cyclic di nucleotide phoshodiesterase | | Authors: | Yadav, M, Pal, K, Sen, U. | | Deposit date: | 2018-09-28 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of c-di-GMP/cGAMP degrading phosphodiesterase VcEAL: identification of a novel conformational switch and its implication.
Biochem.J., 476, 2019
|
|
7MWB
 
 | | ERAP1 binds peptide C-terminus of a SPF sequence (FKARKF) | | Descriptor: | Endoplasmic reticulum aminopeptidase 1,SPF Sequence | | Authors: | Guo, H.C, Sui, L. | | Deposit date: | 2021-05-16 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | ERAP1 binds peptide C-termini of different sequences and/or lengths by a common recognition mechanism.
Immunobiology, 226, 2021
|
|
8B45
 
 | | Structure of CC-Tri with Aib@b,c: CC-Tri-(UbUc)4 | | Descriptor: | 1,2-ETHANEDIOL, CC-Tri-(UbUc)4, SODIUM ION, ... | | Authors: | Kumar, P, Martin, F.J.O, Dawson, W.M, Zieleniewski, F, Woolfson, D.N. | | Deposit date: | 2022-09-19 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of CC-Tri with Aib@b,c: CC-Tri-(UbUc)4
To Be Published
|
|
7R5C
 
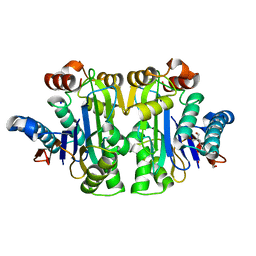 | | Structure of E.coli Class 2 L-asparaginase EcAIII, mutant RDM1-29 (G206C, R207S, D210L, S211V) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Isoaspartyl peptidase, ... | | Authors: | Barciszewski, J, Imiolczyk, B, Loch, J.I, Jaskolski, M. | | Deposit date: | 2022-02-10 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biophysical studies of new L-asparaginase variants: lessons from random mutagenesis of the prototypic Escherichia coli Ntn-amidohydrolase.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6RC4
 
 | |
7M2G
 
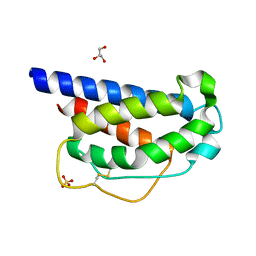 | | INTERLEUKIN-2 (human) mutant P65K, C125S | | Descriptor: | GLYCEROL, Interleukin-2, SULFATE ION | | Authors: | Ptacin, J.L, Milla, M.E, Steinbacher, S, Maskos, K, Thomsen, M. | | Deposit date: | 2021-03-16 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | An engineered IL-2 reprogrammed for anti-tumor therapy using a semi-synthetic organism.
Nat Commun, 12, 2021
|
|
6IFQ
 
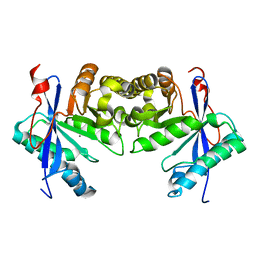 | |
6H4R
 
 | | Crystal structure of human KDM4A in complex with compound 17f | | Descriptor: | 8-[4-[2-[4-[4-[2-(dimethylamino)ethyl]phenyl]piperidin-1-yl]ethyl]pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | C8-substituted pyrido[3,4-d]pyrimidin-4(3H)-ones: Studies towards the identification of potent, cell penetrant Jumonji C domain containing histone lysine demethylase 4 subfamily (KDM4) inhibitors, compound profiling in cell-based target engagement assays.
Eur.J.Med.Chem., 177, 2019
|
|
7R8Q
 
 | | Closed form of SAOUHSC_02373 in complex with ADP, citrate, Mg2+ and Na+ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-grasp domain-containing protein, CITRIC ACID, ... | | Authors: | Pederick, J.L, Bruning, J.B. | | Deposit date: | 2021-06-27 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of an ʟ-amino acid ligase implicated in Staphylococcal sulfur amino acid metabolism.
J.Biol.Chem., 298, 2022
|
|
8BO9
 
 | | NanoLuc-D9R/H57A/K89R mutant complexed with azacoelenterazine bound in intra-barrel catalytic site | | Descriptor: | 3-(4-hydroxyphenyl)-8-[(4-hydroxyphenyl)methyl]-5-(phenylmethyl)-1$l^{4},4,7,8-tetrazabicyclo[4.3.0]nona-1(6),2,4-trien-9-one, Non structural polyprotein | | Authors: | Marek, M, Janin, L.Y. | | Deposit date: | 2022-11-15 | | Release date: | 2023-09-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Illuminating the mechanism and allosteric behavior of NanoLuc luciferase.
Nat Commun, 14, 2023
|
|
7MF4
 
 | |
5TXE
 
 | |
6RCK
 
 | | Crystal structure of the OmpK36 GD insertion chimera from Klebsiella pneumonia | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, LAURYL DIMETHYLAMINE-N-OXIDE, ... | | Authors: | Beis, K, Romano, M, Kwong, J. | | Deposit date: | 2019-04-11 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.029 Å) | | Cite: | OmpK36-mediated Carbapenem resistance attenuates ST258 Klebsiella pneumoniae in vivo.
Nat Commun, 10, 2019
|
|
6H4Z
 
 | | Crystal structure of human KDM5B in complex with compound 16a | | Descriptor: | 1,2-ETHANEDIOL, 8-[4-[2-[4-(3-chlorophenyl)piperidin-1-yl]ethyl]pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.V, Velupillai, S, van Montfort, R.L.M. | | Deposit date: | 2018-07-23 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | C8-substituted pyrido[3,4-d]pyrimidin-4(3H)-ones: Studies towards the identification of potent, cell penetrant Jumonji C domain containing histone lysine demethylase 4 subfamily (KDM4) inhibitors, compound profiling in cell-based target engagement assays.
Eur.J.Med.Chem., 177, 2019
|
|
8BWG
 
 | | HRas (1-166) Y64 phosphorylation | | Descriptor: | GTPase HRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Baumann, P, Jin, Y. | | Deposit date: | 2022-12-06 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Far-reaching effects of tyrosine64 phosphorylation on Ras revealed with BeF 3 - complexes.
Commun Chem, 7, 2024
|
|
7MJU
 
 | |
7R2W
 
 | | Mutant S-adenosylmethionine synthetase from E.coli complexed with AMPPNP and methionine | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Shahar, A, Kleiner, D, Bershtein, S, Zarivach, R. | | Deposit date: | 2022-02-06 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evolution of homo-oligomerization of methionine S-adenosyltransferases is replete with structure-function constrains.
Protein Sci., 31, 2022
|
|
