2NTZ
 
 | |
1V9T
 
 | | Structure of E. coli cyclophilin B K163T mutant bound to succinyl-ALA-PRO-ALA-P-nitroanilide | | Descriptor: | (SIN)APA(NIT), cyclophilin B | | Authors: | Konno, M, Sano, Y, Okudaira, K, Kawaguchi, Y, Yamagishi-Ohmori, Y, Fushinobu, S, Matsuzawa, H. | | Deposit date: | 2004-02-03 | | Release date: | 2004-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Escherichia coli cyclophilin B binds a highly distorted form of trans-prolyl peptide isomer
Eur.J.Biochem., 271, 2004
|
|
2ORZ
 
 | | Structural Basis for Ligand Binding and Heparin Mediated Activation of Neuropilin | | Descriptor: | Neuropilin-1, Tuftsin | | Authors: | Vander Kooi, C.W, Jusino, M.A, Perman, B, Neau, D.B, Bellamy, H.D, Leahy, D.J. | | Deposit date: | 2007-02-05 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for ligand and heparin binding to neuropilin B domains.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3TWI
 
 | | Variable Lymphocyte Receptor Recognition of the Immunodominant Glycoprotein of Bacillus anthracis Spores | | Descriptor: | BclA protein, GLYCEROL, Variable lymphocyte receptor B | | Authors: | Kirchdoerfer, R.N, Herrin, B.R, Han, B.W, Turnbough Jr, C.L, Cooper, M.D, Wilson, I.A. | | Deposit date: | 2011-09-21 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Variable Lymphocyte Receptor Recognition of the Immunodominant Glycoprotein of Bacillus anthracis Spores.
Structure, 20, 2012
|
|
1DV1
 
 | | STRUCTURE OF BIOTIN CARBOXYLASE (APO) | | Descriptor: | BIOTIN CARBOXYLASE, PHOSPHATE ION | | Authors: | Thoden, J.B, Blanchard, C.Z, Holden, H.M, Waldrop, G.L. | | Deposit date: | 2000-01-19 | | Release date: | 2000-06-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Movement of the biotin carboxylase B-domain as a result of ATP binding.
J.Biol.Chem., 275, 2000
|
|
3ZTA
 
 | | The bacterial stressosome: a modular system that has been adapted to control secondary messenger signaling | | Descriptor: | ANTI-SIGMA-FACTOR ANTAGONIST (STAS) DOMAIN PROTEIN | | Authors: | Quin, M.B, Berrisford, J.M, Newman, J.A, Basle, A, Lewis, R.J, Marles-Wright, J. | | Deposit date: | 2011-07-06 | | Release date: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Bacterial Stressosome: A Modular System that Has Been Adapted to Control Secondary Messenger Signaling.
Structure, 20, 2012
|
|
6MHN
 
 | | Photoactive Yellow Protein with covalently bound 3-chloro-4-hydroxycinnamic acid chromophore | | Descriptor: | (2E)-3-(3-chloro-4-hydroxyphenyl)prop-2-enoic acid, Photoactive yellow protein | | Authors: | Thomson, B.D, Both, J, Wu, Y, Parrish, R.M, Martinez, T, Boxer, S.G. | | Deposit date: | 2018-09-18 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Perturbation of Short Hydrogen Bonds in Photoactive Yellow Protein via Noncanonical Amino Acid Incorporation.
J.Phys.Chem.B, 123, 2019
|
|
2AQX
 
 | | Crystal Structure of the Catalytic and CaM-Binding domains of Inositol 1,4,5-Trisphosphate 3-Kinase B | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PREDICTED: inositol 1,4,5-trisphosphate 3-kinase B | | Authors: | Chamberlain, P.P, Sandberg, M.L, Sauer, K, Cooke, M.P, Lesley, S.A, Spraggon, G. | | Deposit date: | 2005-08-18 | | Release date: | 2005-12-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into enzyme regulation for inositol 1,4,5-trisphosphate 3-kinase B
Biochemistry, 44, 2005
|
|
1Q5D
 
 | | Epothilone B-bound Cytochrome P450epoK | | Descriptor: | 7,11-DIHYDROXY-8,8,10,12,16-PENTAMETHYL-3-[1-METHYL-2-(2-METHYL-THIAZOL-4-YL)VINYL]-4,17-DIOXABICYCLO[14.1.0]HEPTADECANE-5,9-DIONE, P450 epoxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagano, S, Li, H, Shimizu, H, Nishida, C, Ogura, H, Ortiz de Montellano, P.R, Poulos, T.L. | | Deposit date: | 2003-08-06 | | Release date: | 2003-10-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of epothilone D-bound, epothilone B-bound, and substrate-free forms of cytochrome P450epoK
J.Biol.Chem., 278, 2003
|
|
2B3D
 
 | |
1DSL
 
 | | GAMMA B CRYSTALLIN C-TERMINAL DOMAIN | | Descriptor: | GAMMA B CRYSTALLIN | | Authors: | Norledge, B.V, Mayr, E.-M, Glockshuber, R, Bateman, O.A, Slingsby, C, Jaenicke, R, Driessen, H.P.C. | | Deposit date: | 1996-02-01 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The X-ray structures of two mutant crystallin domains shed light on the evolution of multi-domain proteins.
Nat.Struct.Biol., 3, 1996
|
|
1DLE
 
 | | FACTOR B SERINE PROTEASE DOMAIN | | Descriptor: | COMPLEMENT FACTOR B | | Authors: | Jing, H, Xu, Y, Carson, M, Moore, D, Macon, K.J, Volanakis, J.E, Narayana, S.V. | | Deposit date: | 1999-12-09 | | Release date: | 2000-12-13 | | Last modified: | 2019-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New structural motifs on the chymotrypsin fold and their potential roles in complement factor B.
EMBO J., 19, 2000
|
|
6LZB
 
 | | crystal structure of Human Methionine aminopeptidase (HsMetAP1b) in complex with AN-P2-5H-06 | | Descriptor: | 1-[(3-methoxyphenyl)methyl]-~{N}-oxidanyl-pyrrolo[2,3-b]pyridine-5-carboxamide, COBALT (II) ION, GLYCEROL, ... | | Authors: | sandeep, C.B, Addlagatta, A. | | Deposit date: | 2020-02-18 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Selective inhibition of Helicobacter pylori methionine aminopeptidase by azaindole hydroxamic acid derivatives: Design, synthesis, in vitro biochemical and structural studies.
Bioorg.Chem., 115, 2021
|
|
6LZC
 
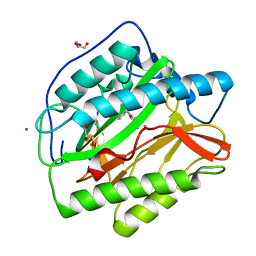 | | crystal structure of Human Methionine aminopeptidase (HsMetAP1b) in complex with KV-P2-4H-05 | | Descriptor: | COBALT (II) ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Sandeep, C.B, Addlagatta, A. | | Deposit date: | 2020-02-18 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Selective inhibition of Helicobacter pylori methionine aminopeptidase by azaindole hydroxamic acid derivatives: Design, synthesis, in vitro biochemical and structural studies.
Bioorg.Chem., 115, 2021
|
|
6MHI
 
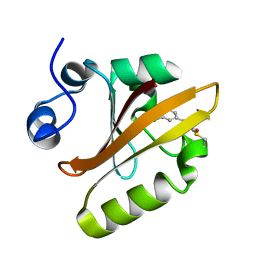 | | Photoactive Yellow Protein with covalently bound 3,5-dichloro-4-hydroxycinnamic acid chromophore | | Descriptor: | (2E)-3-(3,5-dichloro-4-hydroxyphenyl)prop-2-enoic acid, Photoactive yellow protein | | Authors: | Thomson, B.D, Both, J, Wu, Y, Parrish, R.M, Martinez, T, Boxer, S.G. | | Deposit date: | 2018-09-18 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Perturbation of Short Hydrogen Bonds in Photoactive Yellow Protein via Noncanonical Amino Acid Incorporation.
J.Phys.Chem.B, 123, 2019
|
|
1MQZ
 
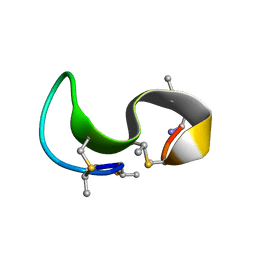 | | NMR solution structure of type-B lantibiotics mersacidin bound to lipid II in DPC micelles | | Descriptor: | LANTIBIOTIC MERSACIDIN | | Authors: | Hsu, S.-T, Breukink, E, Bierbaum, G, Sahl, H.-G, de Kruijff, B, Kaptein, R, van Nuland, N.A, Bonvin, A.M. | | Deposit date: | 2002-09-17 | | Release date: | 2003-03-11 | | Last modified: | 2024-07-10 | | Method: | SOLUTION NMR | | Cite: | NMR Study of Mersacidin and Lipid II Interaction in Dodecylphosphocholine Micelles. Conformational Changes are a Key to Antimicrobial Activity
J.Biol.Chem., 278, 2003
|
|
4FLU
 
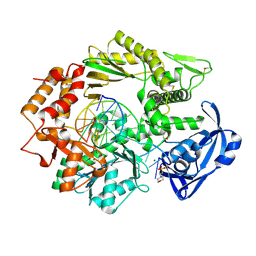 | |
1MQY
 
 | | NMR solution structure of type-B lantibiotics mersacidin in DPC micelles | | Descriptor: | LANTIBIOTIC MERSACIDIN | | Authors: | Hsu, S.-T, Breukink, E, Bierbaum, G, Sahl, H.-G, de Kruijff, B, Kaptein, R, van Nuland, N.A, Bonvin, A.M. | | Deposit date: | 2002-09-17 | | Release date: | 2003-03-11 | | Last modified: | 2024-07-10 | | Method: | SOLUTION NMR | | Cite: | NMR Study of Mersacidin and Lipid II Interaction in Dodecylphosphocholine Micelles. Conformational Changes are a Key to Antimicrobial Activity
J.Biol.Chem., 278, 2003
|
|
2AMJ
 
 | |
6MKT
 
 | | Photoactive Yellow Protein with 3-chlorotyrosine substituted at position 42 | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Thomson, B.D, Both, J, Wu, Y, Parrish, R.M, Martinez, T, Boxer, S.G. | | Deposit date: | 2018-09-26 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Perturbation of Short Hydrogen Bonds in Photoactive Yellow Protein via Noncanonical Amino Acid Incorporation.
J.Phys.Chem.B, 123, 2019
|
|
3MB6
 
 | | Human CK2 catalytic domain in complex with a difurane derivative inhibitor (CPA) | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION, naphtho[2,1-b:7,6-b']difuran-2,8-dicarboxylic acid | | Authors: | Reiser, J.-B, Prudent, R, Claude, C. | | Deposit date: | 2010-03-25 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | New potent dual inhibitors of CK2 and Pim kinases: discovery and structural insights.
Faseb J., 24, 2010
|
|
3HCI
 
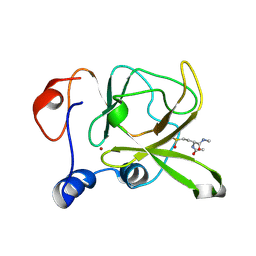 | | Structure of MsrB from Xanthomonas campestris (complex-like form) | | Descriptor: | (2S)-2-(acetylamino)-N-methyl-4-[(R)-methylsulfinyl]butanamide, CALCIUM ION, Peptide methionine sulfoxide reductase, ... | | Authors: | Ranaivoson, F.M, Kauffmann, B, Favier, F. | | Deposit date: | 2009-05-06 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Methionine sulfoxide reductase B displays a high level of flexibility.
J.Mol.Biol., 394, 2009
|
|
3HCV
 
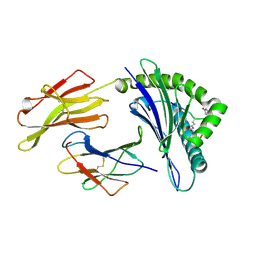 | | Crystal structure of HLA-B*2709 complexed with the double citrullinated vasoactive intestinal peptide type 1 receptor (VIPR) peptide (residues 400-408) | | Descriptor: | Beta-2-microglobulin, DOUBLE CITRULLINATED VASOACTIVE INTESTINAL POLYPEPTIDE RECEPTOR, HLA class I histocompatibility antigen, ... | | Authors: | Beltrami, A, Gabdulkhakov, A, Rossmann, M, Ziegler, A, Uchanska-Ziegler, B, Saenger, W. | | Deposit date: | 2009-05-06 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Citrullination-and mhc polymorphism-dependent conformational changes of
a self peptide
To be Published
|
|
3HCJ
 
 | |
1SQV
 
 | | Crystal Structure Analysis of Bovine Bc1 with UHDBT | | Descriptor: | 6-HYDROXY-5-UNDECYL-1,3-BENZOTHIAZOLE-4,7-DIONE, Cytochrome b, Cytochrome c1, ... | | Authors: | Esser, L, Quinn, B, Li, Y.F, Zhang, M, Elberry, M, Yu, L, Yu, C.A, Xia, D. | | Deposit date: | 2004-03-19 | | Release date: | 2005-09-06 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystallographic studies of quinol oxidation site inhibitors: a modified classification of inhibitors for the cytochrome bc(1) complex.
J.Mol.Biol., 341, 2004
|
|
