2G7I
 
 | |
6MP6
 
 | | Cryo-EM structure of the human neutral amino acid transporter ASCT2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neutral amino acid transporter B(0) | | Authors: | Yu, X, Han, S. | | Deposit date: | 2018-10-05 | | Release date: | 2019-11-06 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Cryo-EM structures of the human glutamine transporter SLC1A5 (ASCT2) in the outward-facing conformation.
Elife, 8, 2019
|
|
6MJA
 
 | | Crystal structure of the mCD1d/xxo (JJ294) /iNKTCR ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Bitra, A, Janssens, J. | | Deposit date: | 2018-09-20 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 4"-O-Alkylated alpha-Galactosylceramide Analogues as iNKT-Cell Antigens: Synthetic, Biological, and Structural Studies.
ChemMedChem, 14, 2019
|
|
3DC9
 
 | | Use of Carbonic Anhydrase II, IX Active-Site Mimic, for the Purpose of Screening Inhibitors for Possible Anti-Cancer Properties | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Genis, C, Sippel, K.H, Case, N, Govindasamy, L, Agbandje-Mckenna, M, Mckenna, R. | | Deposit date: | 2008-06-03 | | Release date: | 2009-03-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design of a carbonic anhydrase IX active-site mimic to screen inhibitors for possible anticancer properties
Biochemistry, 48, 2009
|
|
6MJQ
 
 | | Crystal structure of the mCD1d/xxp (JJ295) /iNKTCR ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Bitra, A, Janssens, J. | | Deposit date: | 2018-09-21 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 4"-O-Alkylated alpha-Galactosylceramide Analogues as iNKT-Cell Antigens: Synthetic, Biological, and Structural Studies.
ChemMedChem, 14, 2019
|
|
3LU6
 
 | | Human serum albumin in complex with compound 1 | | Descriptor: | Serum albumin, [(1R,2R)-2-{[(5-fluoro-1H-indol-2-yl)carbonyl]amino}-2,3-dihydro-1H-inden-1-yl]acetic acid | | Authors: | Buttar, D, Colclough, N, Gerhardt, S, MacFaul, P.A, Phillips, S.D, Plowright, A, Whittamore, P, Tam, K, Maskos, K, Steinbacher, S, Steuber, H. | | Deposit date: | 2010-02-17 | | Release date: | 2010-10-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A combined spectroscopic and crystallographic approach to probing drug-human serum albumin interactions
Bioorg.Med.Chem., 18, 2010
|
|
6MJI
 
 | | Crystal structure of the mCD1d/xxs (JJ304) /iNKTCR ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Bitra, A, Janssens, J. | | Deposit date: | 2018-09-20 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 4"-O-Alkylated alpha-Galactosylceramide Analogues as iNKT-Cell Antigens: Synthetic, Biological, and Structural Studies.
ChemMedChem, 14, 2019
|
|
1E36
 
 | | PORCINE PANCREATIC ELASTASE COMPLEXED WITH (3S, 4S)N-PARA-NITROBENZENESULPHONYL -3-ETHYL-4-(CARBOXYLIC ACID)PYRROLIDIN-2-ONE | | Descriptor: | (2S,3S)-3-FORMYL-2-({[(4-NITROPHENYL)SULFONYL]AMINO}METHYL)PENTANOIC ACID, CALCIUM ION, ELASTASE, ... | | Authors: | Wright, P.A, Wilmouth, R.C, Clifton, I.J, Schofield, C.J. | | Deposit date: | 2000-06-06 | | Release date: | 2000-10-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 'Ph-Jump' Crystallographic Analyses of Gamma-Lactam-Porcine Pancreatic Elastase Complexes
Biochem.J., 351, 2000
|
|
3DBU
 
 | | Use of Carbonic Anhydrase II, IX Active-Site Mimic, for the Purpose of Screening Inhibitors for Possible Anti-Cancer Properties | | Descriptor: | 5-[(phenylsulfonyl)amino]-1,3,4-thiadiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Genis, C, Sippel, K.H, Case, N, Govindasamy, L, Agbandje-Mckenna, M, Mckenna, R. | | Deposit date: | 2008-06-02 | | Release date: | 2009-03-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design of a carbonic anhydrase IX active-site mimic to screen inhibitors for possible anticancer properties
Biochemistry, 48, 2009
|
|
1GJF
 
 | | Peptide Antagonist of IGFBP1, (i,i+7) Covalently Restrained Analog, Minimized Average Structure | | Descriptor: | IGFBP-1 antagonist, PENTANE | | Authors: | Skelton, N.J, Chen, Y.M, Dubree, N, Quan, C, Jackson, D.Y, Cochran, A.G, Zobel, K, Deshayes, K, Baca, M, Pisabarro, M.T, Lowman, H.B. | | Deposit date: | 2001-05-11 | | Release date: | 2001-05-30 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of a phage display-derived peptide that binds to insulin-like growth factor binding protein 1.
Biochemistry, 40, 2001
|
|
3DD0
 
 | | Use of Carbonic Anhydrase II, IX Active-Site Mimic, for the Purpose of Screening Inhibitors for Possible Anti-Cancer Properties | | Descriptor: | 6-ethoxy-1,3-benzothiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Genis, C, Sippel, K.H, Case, N, Govindasamy, L, Agbandje-Mckenna, M, Mckenna, R. | | Deposit date: | 2008-06-04 | | Release date: | 2009-03-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Design of a carbonic anhydrase IX active-site mimic to screen inhibitors for possible anticancer properties
Biochemistry, 48, 2009
|
|
2R2G
 
 | | Structure of Eugenol Synthase from Ocimum basilicum complexed with EMDF | | Descriptor: | Eugenol synthase 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ethyl (1S,2S)-2-(4-hydroxy-3-methoxyphenyl)cyclopropanecarboxylate | | Authors: | Louie, G.V, Noel, J.P, Bowman, M.E. | | Deposit date: | 2007-08-24 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and reaction mechanism of basil eugenol synthase
Plos One, 2, 2007
|
|
6M1X
 
 | |
6MU4
 
 | | Bst DNA polymerase I FANA/DNA binary complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(P*GP*CP*GP*AP*TP*CP*AP*CP*GP*T)-3'), DNA polymerase I, ... | | Authors: | Jackson, L.N, Chim, N, Chaput, J.C. | | Deposit date: | 2018-10-22 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structures of a natural DNA polymerase that functions as an XNA reverse transcriptase.
Nucleic Acids Res., 47, 2019
|
|
1MZK
 
 | | NMR structure of kinase-interacting FHA domain of kinase associated protein phosphatase, KAPP in Arabidopsis | | Descriptor: | KINASE ASSOCIATED PROTEIN PHOSPHATASE | | Authors: | Lee, G, Ding, Z, Walker, J.C, Van Doren, S.R. | | Deposit date: | 2002-10-08 | | Release date: | 2003-09-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the forkhead-associated domain from the Arabidopsis receptor kinase-associated protein phosphatase.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
6MUP
 
 | |
6MIY
 
 | | Crystal structure of the mCD1d/xxa (JJ239)/iNKTCR ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Bitra, A, Janssens, J. | | Deposit date: | 2018-09-20 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | 4"-O-Alkylated alpha-Galactosylceramide Analogues as iNKT-Cell Antigens: Synthetic, Biological, and Structural Studies.
ChemMedChem, 14, 2019
|
|
6MJJ
 
 | | Crystal structure of the mCD1d/xxm (JJ290) /iNKTCR ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zajonc, D.M, Bitra, A, Janssens, J. | | Deposit date: | 2018-09-21 | | Release date: | 2019-01-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | 4"-O-Alkylated alpha-Galactosylceramide Analogues as iNKT-Cell Antigens: Synthetic, Biological, and Structural Studies.
ChemMedChem, 14, 2019
|
|
2G86
 
 | |
2G8D
 
 | |
2G89
 
 | |
1E35
 
 | | PORCINE PANCREATIC ELASTASE COMPLEXED WITH (3S, 4S)N-PARA-TOLUENESULPHONYL -3-ETHYL-4-(CARBOXYLIC ACID)PYRROLIDIN-2-ONE SOAKED IN PH 9 BUFFER FOR TWO MINUTES | | Descriptor: | (2S,3S)-3-FORMYL-2-({[(4-METHYLPHENYL)SULFONYL]AMINO}METHYL)PENTANOIC ACID, CALCIUM ION, ELASTASE, ... | | Authors: | Wright, P.A, Wilmouth, R.C, Clifton, I.J, Schofield, C.J. | | Deposit date: | 2000-06-06 | | Release date: | 2000-10-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 'Ph-Jump' Crystallographic Analyses of Gamma-Lactam-Porcine Pancreatic Elastase Complexes
Biochem.J., 351, 2000
|
|
2QYS
 
 | |
2QYF
 
 | |
2KSQ
 
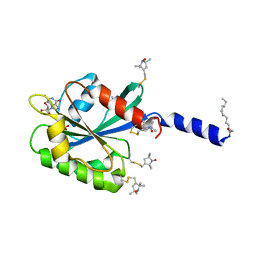 | | The myristoylated yeast ARF1 in a GTP and bicelle bound conformation | | Descriptor: | ADP-ribosylation factor 1, GUANOSINE-5'-TRIPHOSPHATE, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Liu, Y, Kahn, R, Prestegard, J. | | Deposit date: | 2010-01-12 | | Release date: | 2010-07-07 | | Last modified: | 2011-07-27 | | Method: | SOLUTION NMR | | Cite: | Dynamic structure of membrane-anchored Arf*GTP.
Nat.Struct.Mol.Biol., 17, 2010
|
|
