3PVD
 
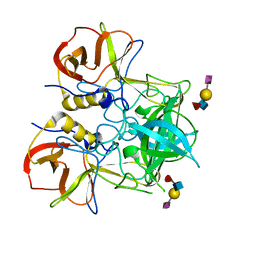 | | Crystal structure of P domain dimer of Norovirus VA207 complexed with 3'-sialyl-Lewis x tetrasaccharide | | Descriptor: | Capsid, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Chen, Y, Tan, M, Xia, M, Hao, N, Zhang, X.C, Huang, P, Jiang, X, Li, X, Rao, Z. | | Deposit date: | 2010-12-06 | | Release date: | 2011-08-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallography of a Lewis-binding norovirus, elucidation of strain-specificity to the polymorphic human histo-blood group antigens
Plos Pathog., 7, 2011
|
|
3PVM
 
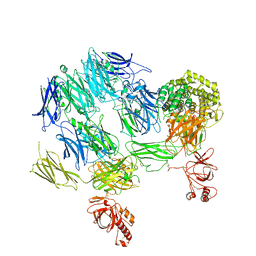 | | Structure of Complement C5 in Complex with CVF | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cobra venom factor, Complement C5 | | Authors: | Laursen, N.S, Andersen, K.R, Braren, I, Sottrup-Jensen, L, Spillner, E, Andersen, G.R. | | Deposit date: | 2010-12-07 | | Release date: | 2011-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Substrate recognition by complement convertases revealed in the C5-cobra venom factor complex.
Embo J., 30, 2011
|
|
1I6Z
 
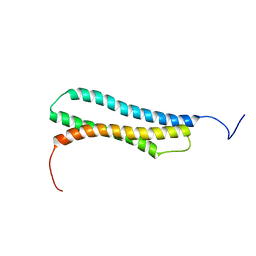 | | BAG DOMAIN OF BAG1 COCHAPERONE | | Descriptor: | BAG-FAMILY MOLECULAR CHAPERONE REGULATOR-1 | | Authors: | Briknarova, K, Takayama, S, Brive, L, Havert, M.L, Knee, D.A, Velasco, J, Homma, S, Cabezas, E, Stuart, J, Hoyt, D.W, Satterthwait, A.C, Llinas, M, Reed, J.C, Ely, K.R. | | Deposit date: | 2001-03-06 | | Release date: | 2001-09-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of BAG1 cochaperone and its interactions with Hsc70 heat shock protein.
Nat.Struct.Biol., 8, 2001
|
|
1I8N
 
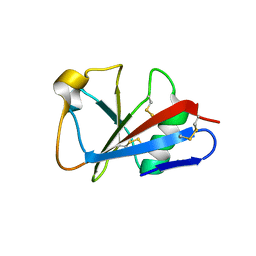 | | CRYSTAL STRUCTURE OF LEECH ANTI-PLATELET PROTEIN | | Descriptor: | ANTI-PLATELET PROTEIN, PROPIONAMIDE | | Authors: | Huizinga, E.G, Schouten, A, Connolly, T.M, Kroon, J, Sixma, J.J, Gros, P. | | Deposit date: | 2001-03-15 | | Release date: | 2001-08-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of leech anti-platelet protein, an inhibitor of haemostasis.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
7N14
 
 | | Crystal structure of 4-(1H-1,2,4-triazol-1-yl)benzoic acid-bound CYP199A4 | | Descriptor: | 4-(1H-1,2,4-triazol-1-yl)benzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Podgorski, M.N, Bruning, J.B, Bell, S.G. | | Deposit date: | 2021-05-26 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.537 Å) | | Cite: | To Be, or Not to Be, an Inhibitor: A Comparison of Azole Interactions with and Oxidation by a Cytochrome P450 Enzyme.
Inorg.Chem., 61, 2022
|
|
5VRI
 
 | | Crystal structure of SsoPox AsA6 mutant (F46L-C258A-W263M-I280T) - closed form | | Descriptor: | 1,2-ETHANEDIOL, Aryldialkylphosphatase, COBALT (II) ION, ... | | Authors: | Hiblot, J, Gotthard, G, Jacquet, P, Daude, D, Bergonzi, C, Chabriere, E, Elias, M. | | Deposit date: | 2017-05-10 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Rational engineering of a native hyperthermostable lactonase into a broad spectrum phosphotriesterase.
Sci Rep, 7, 2017
|
|
1B89
 
 | | CLATHRIN HEAVY CHAIN PROXIMAL LEG SEGMENT (BOVINE) | | Descriptor: | PROTEIN (CLATHRIN HEAVY CHAIN) | | Authors: | Ybe, J.A, Brodsky, F.M, Hofmann, K, Lin, K, Liu, S.-H, Chen, L, Earnest, T.N, Fletterick, R.J, Hwang, P.K. | | Deposit date: | 1999-05-27 | | Release date: | 1999-06-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Clathrin self-assembly is mediated by a tandemly repeated superhelix.
Nature, 399, 1999
|
|
1I9H
 
 | | CORE STREPTAVIDIN-BNA COMPLEX | | Descriptor: | 5-(2-OXO-HEXAHYDRO-THIENO[3,4-D]IMIDAZOL-6-YL)-PENTANOIC ACID (4-NITRO-PHENYL)-AMIDE, STREPTAVIDIN | | Authors: | Livnah, O, Huberman, T, Wilchek, M, Bayer, E.A, Eisenberg-Domovich, Y. | | Deposit date: | 2001-03-20 | | Release date: | 2001-09-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Chicken avidin exhibits pseudo-catalytic properties. Biochemical, structural, and electrostatic consequences.
J.Biol.Chem., 276, 2001
|
|
1B9N
 
 | | REGULATOR FROM ESCHERICHIA COLI | | Descriptor: | NICKEL (II) ION, PROTEIN (MODE) | | Authors: | Hall, D.R, Gourley, D.G, Hunter, W.N. | | Deposit date: | 1999-02-12 | | Release date: | 2000-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The high-resolution crystal structure of the molybdate-dependent transcriptional regulator (ModE) from Escherichia coli: a novel combination of domain folds.
EMBO J., 18, 1999
|
|
3BN1
 
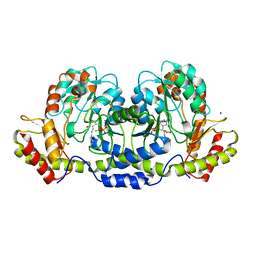 | | Crystal structure of GDP-perosamine synthase | | Descriptor: | 2-OXOGLUTARIC ACID, ACETATE ION, Perosamine synthetase, ... | | Authors: | Cook, P.D, Holden, H.M. | | Deposit date: | 2007-12-13 | | Release date: | 2008-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | GDP-Perosamine Synthase: Structural Analysis and Production of a Novel Trideoxysugar
Biochemistry, 47, 2008
|
|
1B9M
 
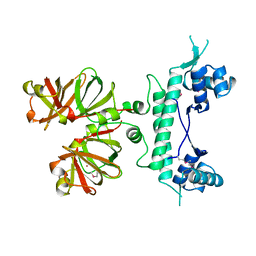 | | REGULATOR FROM ESCHERICHIA COLI | | Descriptor: | NICKEL (II) ION, PROTEIN (MODE) | | Authors: | Hall, D.R, Gourley, D.G, Hunter, W.N. | | Deposit date: | 1999-02-12 | | Release date: | 2000-03-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The high-resolution crystal structure of the molybdate-dependent transcriptional regulator (ModE) from Escherichia coli: a novel combination of domain folds.
EMBO J., 18, 1999
|
|
7N40
 
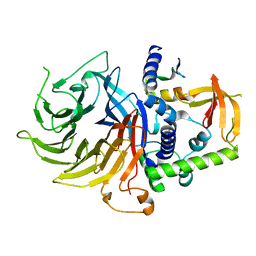 | | Crystal structure of LIN9-RbAp48-LIN37, a MuvB subcomplex | | Descriptor: | Histone-binding protein RBBP4, Isoform 2 of Protein lin-9 homolog, Protein lin-37 homolog | | Authors: | Asthana, A, Ramanan, P, Tripathi, S.M, Rubin, S.M. | | Deposit date: | 2021-06-02 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The MuvB complex binds and stabilizes nucleosomes downstream of the transcription start site of cell-cycle dependent genes.
Nat Commun, 13, 2022
|
|
3PY7
 
 | |
7N3L
 
 | | Co-complex CYP46A1 with 0420 (compound 6) | | Descriptor: | 1,2-ETHANEDIOL, Cholesterol 24-hydroxylase, GLYCEROL, ... | | Authors: | Lane, W, Yano, J. | | Deposit date: | 2021-06-01 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Discovery of Novel 3-Piperidinyl Pyridine Derivatives as Highly Potent and Selective Cholesterol 24-Hydroxylase (CH24H) Inhibitors.
J.Med.Chem., 65, 2022
|
|
1BC7
 
 | | SERUM RESPONSE FACTOR ACCESSORY PROTEIN 1A (SAP-1)/DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*AP*CP*AP*TP*CP*CP*TP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*AP*GP*GP*AP*TP*GP*TP*G)-3'), PROTEIN (ETS-DOMAIN PROTEIN) | | Authors: | Mo, Y, Vaessen, B, Johnston, K, Marmorstein, R. | | Deposit date: | 1998-05-05 | | Release date: | 1999-01-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structures of SAP-1 bound to DNA targets from the E74 and c-fos promoters: insights into DNA sequence discrimination by Ets proteins.
Mol.Cell, 2, 1998
|
|
7N3M
 
 | | Co-complex CYP46A1 with 0431 (compound 17) | | Descriptor: | Cholesterol 24-hydroxylase, N,N-dimethyl-1-[4-(4-methyl-1H-pyrazol-1-yl)pyridin-3-yl]piperidine-4-carboxamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lane, W, Yano, J. | | Deposit date: | 2021-06-01 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Discovery of Novel 3-Piperidinyl Pyridine Derivatives as Highly Potent and Selective Cholesterol 24-Hydroxylase (CH24H) Inhibitors.
J.Med.Chem., 65, 2022
|
|
1AO8
 
 | | DIHYDROFOLATE REDUCTASE COMPLEXED WITH METHOTREXATE, NMR, 21 STRUCTURES | | Descriptor: | DIHYDROFOLATE REDUCTASE, METHOTREXATE | | Authors: | Gargaro, A.R, Soteriou, A, Frenkiel, T.A, Bauer, C.J, Birdsall, B, Polshakov, V.I, Barsukov, I.L, Roberts, G.C.K, Feeney, J. | | Deposit date: | 1997-07-22 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the complex of Lactobacillus casei dihydrofolate reductase with methotrexate.
J.Mol.Biol., 277, 1998
|
|
4IK4
 
 | | High resolution structure of GCaMP3 at pH 5.0 | | Descriptor: | CALCIUM ION, RCaMP, Green fluorescent protein | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-02-05 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
5W48
 
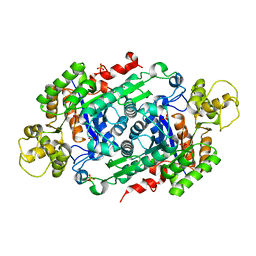 | | Crystal Structure of Riboflavin Lyase (RcaE) | | Descriptor: | Riboflavin Lyase, SULFATE ION | | Authors: | Bhandari, D.M, Chakrabarty, Y, Zhao, B, Wood, J, Li, P, Begley, T.P. | | Deposit date: | 2017-06-09 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cannibalism Among the Flavins: a Novel C-N Bond Cleavage in Riboflavin Catabolism Mediated by Flavin-Generated Superoxide Radical
To be Published
|
|
1APF
 
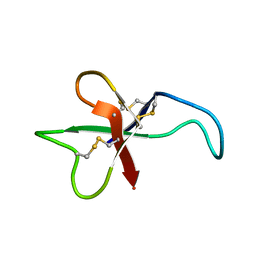 | | ANTHOPLEURIN-B, NMR, 20 STRUCTURES | | Descriptor: | ANTHOPLEURIN-B | | Authors: | Monks, S.A, Pallaghy, P.K, Scanlon, M.J, Norton, R.S. | | Deposit date: | 1995-05-30 | | Release date: | 1996-07-11 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cardiostimulant polypeptide anthopleurin-B and comparison with anthopleurin-A.
Structure, 3, 1995
|
|
5W4X
 
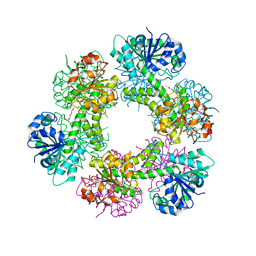 | | Truncated hUGDH | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, UDP-glucose 6-dehydrogenase | | Authors: | Sennett, N.C, Custer, G.S, Wood, Z.A. | | Deposit date: | 2017-06-13 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The entropic force generated by intrinsically disordered segments tunes protein function.
Nature, 563, 2018
|
|
3BSZ
 
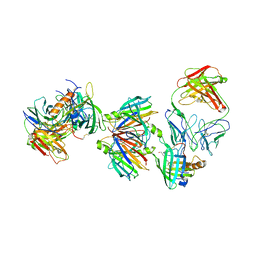 | | Crystal structure of the transthyretin-retinol binding protein-Fab complex | | Descriptor: | Fab fragment heavy chain, Fab fragment light chain, Plasma retinol-binding protein, ... | | Authors: | Zanotti, G, Cendron, L, Gliubich, F, Folli, C, Berni, R. | | Deposit date: | 2007-12-27 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Structural and mutational analyses of protein-protein interactions between transthyretin and retinol-binding protein.
Febs J., 275, 2008
|
|
4INO
 
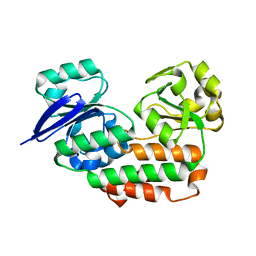 | |
5W4Z
 
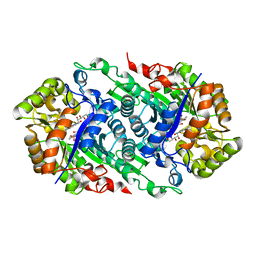 | | Crystal Structure of Riboflavin Lyase (RcaE) with modified FMN and substrate Riboflavin | | Descriptor: | 1-deoxy-1-(7,8-dimethyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl)-3-O-phosphono-D-ribitol, RIBOFLAVIN, Riboflavin Lyase | | Authors: | Bhandari, D.M, Chakrabarty, Y, Zhao, B, Wood, J, Li, P, Begley, T.P. | | Deposit date: | 2017-06-13 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cannibalism Among the Flavins: a Novel C-N Bond Cleavage in Riboflavin Catabolism Mediated by Flavin-Generated Superoxide Radical
To be Published
|
|
1W6T
 
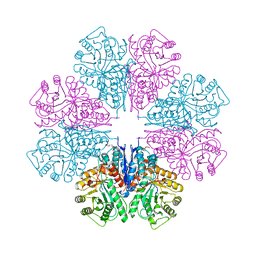 | | Crystal Structure Of Octameric Enolase From Streptococcus pneumoniae | | Descriptor: | ENOLASE, MAGNESIUM ION, NONAETHYLENE GLYCOL | | Authors: | Ehinger, S, Schubert, W.-D, Bergmann, S, Hammerschmidt, S, Heinz, D.W. | | Deposit date: | 2004-08-24 | | Release date: | 2005-08-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Plasmin(Ogen)-Binding Alpha-Enolase from Streptococcus Pneumoniae: Crystal Structure and Evaluation of Plasmin(Ogen)-Binding Sites
J.Mol.Biol., 343, 2004
|
|
