2VW5
 
 | | Structure Of The Hsp90 Inhibitor 7-O-carbamoylpremacbecin Bound To The N- Terminus Of Yeast Hsp90 | | Descriptor: | (4E,8S,9R,10E,12S,13R,14S,16R)-13,20-dihydroxy-14-methoxy-4,8,10,12,16-pentamethyl-3-oxo-2-azabicyclo[16.3.1]docosa-1(22),4,10,18,20-pentaen-9-yl carbamate, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82, SULFATE ION | | Authors: | Roe, S.M, Prodromou, C, Pearl, L.H. | | Deposit date: | 2008-06-16 | | Release date: | 2008-09-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimizing Natural Products by Biosynthetic Engineering: Discovery of Nonquinone Hsp90 Inhibitors.
J.Med.Chem., 51, 2008
|
|
2F37
 
 | |
2VWC
 
 | |
6PIS
 
 | |
9BA5
 
 | |
5B4X
 
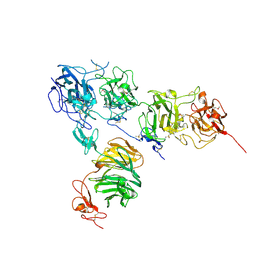 | | Crystal structure of the ApoER2 ectodomain in complex with the Reelin R56 fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Low density lipoprotein receptor-related protein 8, ... | | Authors: | Yasui, N, Hirai, H, Yamashita, K, Takagi, J, Nogi, T. | | Deposit date: | 2016-04-20 | | Release date: | 2017-04-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the ectodomain from a LDLR close homologue in complex with its physiological ligand.
To Be Published
|
|
6R69
 
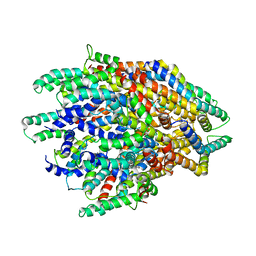 | | Improved map of the FliPQR complex that forms the core of the Salmonella type III secretion system export apparatus. | | Descriptor: | Flagellar biosynthetic protein FliP, Flagellar biosynthetic protein FliQ, Flagellar biosynthetic protein FliR | | Authors: | Johnson, S, Kuhlen, L, Abrusci, P, Lea, S.M. | | Deposit date: | 2019-03-26 | | Release date: | 2019-05-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | The Structure of an Injectisome Export Gate Demonstrates Conservation of Architecture in the Core Export Gate between Flagellar and Virulence Type III Secretion Systems.
Mbio, 10, 2019
|
|
2RMC
 
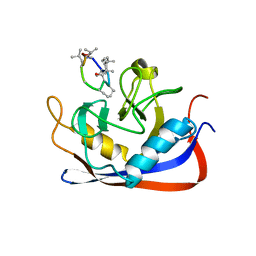 | | Crystal structure of murine cyclophilin C complexed with immunosuppressive drug cyclosporin A | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE C | | Authors: | Ke, H, Zhao, Y, Luo, F, Weissman, I, Friedman, J. | | Deposit date: | 1994-01-07 | | Release date: | 1995-02-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal Structure of Murine Cyclophilin C Complexed with Immunosuppressive Drug Cyclosporin A
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
2RMA
 
 | |
2RMB
 
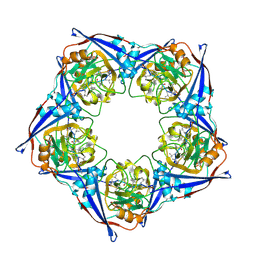 | |
1OHL
 
 | | YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE PUTATIVE CYCLIC REACTION INTERMEDIATE COMPLEX | | Descriptor: | 3-[5-(AMINOMETHYL)-4-(CARBOXYMETHYL)-1H-PYRROL-3-YL]PROPANOIC ACID, BETA-MERCAPTOETHANOL, DELTA-AMINOLEVULINIC ACID DEHYDRATASE, ... | | Authors: | Erskine, P.T, Coates, L, Butler, D, Youell, J.H, Brindley, A.A, Wood, S.P, Warren, M.J, Shoolingin-Jordan, P.M, Cooper, J.B. | | Deposit date: | 2003-05-27 | | Release date: | 2003-06-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-Ray Structure of a Putative Reaction Intermediateof 5-Aminolaevulinic Acid Dehydratase
Biochem.J., 373, 2003
|
|
1VZJ
 
 | | Structure of the tetramerization domain of acetylcholinesterase: four-fold interaction of a WWW motif with a left-handed polyproline helix | | Descriptor: | ACETYLCHOLINESTERASE, ACETYLCHOLINESTERASE COLLAGENIC TAIL PEPTIDE | | Authors: | Dvir, H, Harel, M, Bon, S, Liu, W.-Q, Vidal, M, Garbay, C, Sussman, J.L, Massoulie, J, Silman, I. | | Deposit date: | 2004-05-20 | | Release date: | 2005-01-10 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Synaptic Acetylcholinesterase Tetramer Assembles Around a Polyproline-II Helix
Embo J., 23, 2004
|
|
3UE1
 
 | | Crystal strucuture of Acinetobacter baumanni PBP1A in complex with MC-1 | | Descriptor: | (4S,7Z)-7-(2-amino-1,3-thiazol-4-yl)-1-[({4-[(2R)-2,3-dihydroxypropyl]-3-(4,5-dihydroxypyridin-2-yl)-5-oxo-4,5-dihydro- 1H-1,2,4-triazol-1-yl}sulfonyl)amino]-4-formyl-10,10-dimethyl-1,6-dioxo-9-oxa-2,5,8-triazaundec-7-en-11-oate, Penicillin-binding protein 1a | | Authors: | Han, S. | | Deposit date: | 2011-10-28 | | Release date: | 2011-12-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Distinctive attributes of beta-lactam target proteins in Acinetobacter baumannii relevant to development of new antibiotics
J.Am.Chem.Soc., 133, 2011
|
|
5WE4
 
 | | 70S ribosome-EF-Tu wt complex with GppNHp | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fislage, M, Brown, Z, Frank, J. | | Deposit date: | 2017-07-07 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM shows stages of initial codon selection on the ribosome by aa-tRNA in ternary complex with GTP and the GTPase-deficient EF-TuH84A.
Nucleic Acids Res., 46, 2018
|
|
5WE6
 
 | | 70S ribosome-EF-Tu H84A complex with GTP and cognate tRNA | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fislage, M, Frank, J. | | Deposit date: | 2017-07-07 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM shows stages of initial codon selection on the ribosome by aa-tRNA in ternary complex with GTP and the GTPase-deficient EF-TuH84A.
Nucleic Acids Res., 46, 2018
|
|
5WDT
 
 | | 70S ribosome-EF-Tu H84A complex with GppNHp | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fislage, M, Brown, Z, Frank, J. | | Deposit date: | 2017-07-06 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM shows stages of initial codon selection on the ribosome by aa-tRNA in ternary complex with GTP and the GTPase-deficient EF-TuH84A.
Nucleic Acids Res., 46, 2018
|
|
4UM8
 
 | | Crystal structure of alpha V beta 6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CACODYLATE ION, ... | | Authors: | Dong, X, Springer, T.A. | | Deposit date: | 2014-05-15 | | Release date: | 2014-11-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.852 Å) | | Cite: | Structural Determinants of Integrin Beta-Subunit Specificity for Latent Tgf-Beta
Nat.Struct.Mol.Biol., 21, 2014
|
|
5WZY
 
 | | Crystal structure of the P2X4 receptor from zebrafish in the presence of CTP at 2.8 Angstroms | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYTIDINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Kasuya, G, Hattori, M, Nureki, O. | | Deposit date: | 2017-01-19 | | Release date: | 2017-04-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Structural insights into the nucleotide base specificity of P2X receptors
Sci Rep, 7, 2017
|
|
4UDC
 
 | | GR in complex with dexamethasone | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, DEXAMETHASONE, GLUCOCORTICOID RECEPTOR, ... | | Authors: | Edman, K, Hogner, A, Hussein, A, Bjursell, M, Aagaard, A, Backstrom, S, Bodin, C, Wissler, L, Jellesmark-Jensen, T, Cavallin, A, Karlsson, U, Nilsson, E, Lecina, D, Takahashi, R, Grebner, C, Lepisto, M, Guallar, V. | | Deposit date: | 2014-12-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand Binding Mechanism in Steroid Receptors: From Conserved Plasticity to Differential Evolutionary Constraints.
Structure, 23, 2015
|
|
4UDA
 
 | | MR in complex with dexamethasone | | Descriptor: | DEXAMETHASONE, GLYCEROL, MINERALOCORTICOID RECEPTOR, ... | | Authors: | Edman, K, Hogner, A, Hussein, A, Bjursell, M, Aagaard, A, Backstrom, S, Bodin, C, Wissler, L, Jellesmark-Jensen, T, Cavallin, A, Karlsson, U, Nilsson, E, Lecina, D, Takahashi, R, Grebner, C, Lepisto, M, Guallar, V. | | Deposit date: | 2014-12-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Ligand Binding Mechanism in Steroid Receptors: From Conserved Plasticity to Differential Evolutionary Constraints.
Structure, 23, 2015
|
|
7CJS
 
 | | structure of aquaporin | | Descriptor: | Aquaporin NIP2-1, CHOLESTEROL HEMISUCCINATE, SODIUM ION, ... | | Authors: | Saitoh, Y, Ma, J.F, Suga, M. | | Deposit date: | 2020-07-13 | | Release date: | 2021-11-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for high selectivity of a rice silicon channel Lsi1.
Nat Commun, 12, 2021
|
|
5YDU
 
 | | Crystal structure of Utp30 | | Descriptor: | PHOSPHATE ION, Ribosome biogenesis protein UTP30 | | Authors: | Hu, J, Zhu, X, Ye, K. | | Deposit date: | 2017-09-14 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.646 Å) | | Cite: | Structure and RNA recognition of ribosome assembly factor Utp30.
RNA, 23, 2017
|
|
5YDT
 
 | | Remodeled Utp30 in 90S pre-ribosome (Mtr4-depleted, Enp1-TAP) | | Descriptor: | 5' ETS RNA, Ribosome biogenesis protein UTP30, Saccharomyces cerevisiae strain ALI 308 18S ribosomal RNA gene, ... | | Authors: | Ye, K, Zhu, X, Hu, J. | | Deposit date: | 2017-09-14 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure and RNA recognition of ribosome assembly factor Utp30.
RNA, 23, 2017
|
|
7E36
 
 | | A [6+4]-cycloaddition adduct is the biosynthetic intermediate in streptoseomycin biosynthesis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alkanesulfonate monooxygenase SsuD/methylene tetrahydromethanopterin reductase-like flavin-dependent oxidoreductase (Luciferase family), ... | | Authors: | Zhang, B, Ge, H.M. | | Deposit date: | 2021-02-08 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A [6+4]-cycloaddition adduct is the biosynthetic intermediate in streptoseomycin biosynthesis.
Nat Commun, 12, 2021
|
|
3O83
 
 | | Structure of BasE N-terminal domain from Acinetobacter baumannii bound to 2-(4-n-dodecyl-1,2,3-triazol-1-yl)-5'-O-[N-(2-hydroxybenzoyl)sulfamoyl]adenosine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(4-dodecyl-1H-1,2,3-triazol-1-yl)-5'-O-{[(2-hydroxyphenyl)carbonyl]sulfamoyl}adenosine, ... | | Authors: | Drake, E.J, Duckworth, B.P, Neres, J, Aldrich, C.C, Gulick, A.M. | | Deposit date: | 2010-08-02 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and structural characterization of bisubstrate inhibitors of BasE, the self-standing nonribosomal peptide synthetase adenylate-forming enzyme of acinetobactin synthesis.
Biochemistry, 49, 2010
|
|
