8OX7
 
 | | Cryo-EM structure of ATP8B1-CDC50A in E2P autoinhibited "closed" conformation | | Descriptor: | (2R)-3-{[(R)-{[(1S,2S,3R,4S,5S,6S)-2,6-dihydroxy-3,4,5-tris(phosphonooxy)cyclohexyl]oxy}(hydroxy)phosphoryl]oxy}propane -1,2-diyl dioctanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Cell cycle control protein 50A, ... | | Authors: | Dieudonne, T, Kummerer, F, Juknaviciute Laursen, M, Stock, C, Kock Flygaard, R, Khalid, S, Lenoir, G, Lyons, J.A, Lindorff-Larsen, K, Nissen, P. | | Deposit date: | 2023-05-01 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Activation and substrate specificity of the human P4-ATPase ATP8B1.
Nat Commun, 14, 2023
|
|
8PPN
 
 | |
8PO9
 
 | | Polyethylene oxidation hexamerin PEase Cibeles (XP_026756460) from Galleria mellonella | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Arylphorin, ... | | Authors: | Illanes-Vicioso, R, Ruiz-Lopez, E, Sola, M, Bertocchini, F, Palomo, E.A. | | Deposit date: | 2023-07-03 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Plastic degradation by insect hexamerins: Near-atomic resolution structures of the polyethylene-degrading proteins from the wax worm saliva.
Sci Adv, 9, 2023
|
|
1W3L
 
 | | ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHAERENS IN COMPLEX WITH CELLOTRI DERIVED-TETRAHYDROOXAZINE | | Descriptor: | ENDOGLUCANASE 5A, GLYCEROL, SULFATE ION, ... | | Authors: | Gloster, T.M, Macdonald, J.M, Tarling, C.A, Stick, R.V, Withers, S.W, Davies, G.J. | | Deposit date: | 2004-07-16 | | Release date: | 2004-09-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Structural, Thermodynamic, and Kinetic Analyses of Tetrahydrooxazine-Derived Inhibitors Bound to {Beta}-Glucosidases
J.Biol.Chem., 279, 2004
|
|
8PC0
 
 | | Sub-tomogram average of the open conformation of the Nap adhesion complex from the human pathogen Mycoplasma genitalium. | | Descriptor: | Adhesin P1, Mgp-operon protein 3, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-1,5-anhydro-D-glucitol, ... | | Authors: | Sprankel, L, Scheffer, M.P, Frangakis, A.F. | | Deposit date: | 2023-06-09 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Cryo-electron tomography reveals the binding and release states of the major adhesion complex from Mycoplasma genitalium.
Plos Pathog., 19, 2023
|
|
8PBZ
 
 | |
3HTQ
 
 | | the hemagglutinin structure of an avian H1N1 influenza A virus in complex with LSTc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Wang, G, Li, A, Zhang, Q, Wu, C, Zhang, R, Cai, Q, Song, W, Yuen, K.-Y. | | Deposit date: | 2009-06-12 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | The hemagglutinin structure of an avian H1N1 influenza A virus
Virology, 392, 2009
|
|
5KTI
 
 | | Structure of cow mincle complexed with KMJ1 | | Descriptor: | 2,3-dimethoxybenzoic acid, CALCIUM ION, TRIETHYLENE GLYCOL, ... | | Authors: | Feinberg, H, Rambaruth, N.D.S, Jegouzo, S.A.F, Jacobsen, K.M, Djurhuus, R, Poulsen, T.B, Taylor, M.E, Drickamer, K, Weis, W.I. | | Deposit date: | 2016-07-11 | | Release date: | 2016-08-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding Sites for Acylated Trehalose Analogs of Glycolipid Ligands on an Extended Carbohydrate Recognition Domain of the Macrophage Receptor Mincle.
J.Biol.Chem., 291, 2016
|
|
4M11
 
 | | Crystal Structure of Murine Cyclooxygenase-2 Complex with Meloxicam | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-hydroxy-2-methyl-N-(5-methyl-1,3-thiazol-2-yl)-2H-1,2-benzothiazine-3-carboxamide 1,1-dioxide, ... | | Authors: | Xu, S, Banerjee, S, Hermanson, D.J, Marnett, L.J. | | Deposit date: | 2013-08-02 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Oxicams Bind in a Novel Mode to the Cyclooxygenase Active Site via a Two-water-mediated H-bonding Network.
J.Biol.Chem., 289, 2014
|
|
8PMR
 
 | | NADase from Aspergillus fumigatus with mutated calcium binding motif (D219A/E220A) | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kallio, J.P, Ferrario, E, Stromland, O, Ziegler, M. | | Deposit date: | 2023-06-29 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Novel Calcium-Binding Motif Stabilizes and Increases the Activity of Aspergillus fumigatus Ecto-NADase.
Biochemistry, 62, 2023
|
|
8PMS
 
 | | NADase from Aspergillus fumigatus with replaced C-terminus from Neurospora crassa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kallio, J.P, Ferrario, E, Stromland, O, Ziegler, M. | | Deposit date: | 2023-06-29 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Novel Calcium-Binding Motif Stabilizes and Increases the Activity of Aspergillus fumigatus Ecto-NADase.
Biochemistry, 62, 2023
|
|
5O5U
 
 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with a urea based inhibitor PSMA 1027 | | Descriptor: | (2~{S})-2-[[(2~{S})-6-[[(2~{S})-2-[[4-[[[(2~{S})-2-[[(2~{R})-2-[(6-fluoranylpyridin-3-yl)carbonylamino]-3-sulfo-propanoyl]amino]-3-sulfo-propanoyl]amino]methyl]phenyl]carbonylamino]-3-naphthalen-2-yl-propanoyl]amino]-1-oxidanyl-1-oxidanylidene-hexan-2-yl]carbamoylamino]pentanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Novakova, Z, Barinka, C, Motlova, L. | | Deposit date: | 2017-06-02 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with a urea based inhibitor PSMA 1027
To Be Published
|
|
3L3V
 
 | | Structure of HIV-1 integrase core domain in complex with sucrose | | Descriptor: | CADMIUM ION, POL polyprotein, SULFATE ION, ... | | Authors: | Wielens, J, Chalmers, D.K, Scanlon, M.J, Parker, M.W. | | Deposit date: | 2009-12-18 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the HIV-1 integrase core domain in complex with sucrose reveals details of an allosteric inhibitory binding site
Febs Lett., 584, 2010
|
|
3HT3
 
 | |
6C6A
 
 | | Structure of glycolipid aGSA[16,6P] in complex with mouse CD1d | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M, Wang, J. | | Deposit date: | 2018-01-18 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
8FJC
 
 | | Structure of the catalytic domain of Streptococcus mutans GtfB complexed to acarbose in tetragonal space group P4322 | | Descriptor: | 1,2-ETHANEDIOL, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, CALCIUM ION, ... | | Authors: | Schormann, N, Deivanayagam, C. | | Deposit date: | 2022-12-19 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The catalytic domains of Streptococcus mutans glucosyltransferases: a structural analysis.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
6SHM
 
 | | An inactive (D136A and D137A) variant of alpha-1,6-mannanase, GH76A of Salegentibacter sp. HEL1_6 in complex with alpha-1,6-mannotetrose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Mutant alpha-1,6-mannanase GH76A, ... | | Authors: | Hehemann, J.H, Solanki, V. | | Deposit date: | 2019-08-07 | | Release date: | 2020-09-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Glycoside hydrolase from the GH76 family indicates that marine Salegentibacter sp. Hel_I_6 consumes alpha-mannan from fungi.
Isme J, 2022
|
|
4KRL
 
 | |
3L01
 
 | | Crystal structure of monomeric glycogen synthase from Pyrococcus abyssi | | Descriptor: | CHLORIDE ION, GLYCEROL, GlgA glycogen synthase, ... | | Authors: | Diaz, A, Martinez-Pons, C, Fita, I, Ferrer, J.C, Guinovart, J.J. | | Deposit date: | 2009-12-09 | | Release date: | 2010-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Processivity and Subcellular Localization of Glycogen Synthase Depend on a Non-catalytic High Affinity Glycogen-binding Site.
J.Biol.Chem., 286, 2011
|
|
3LEG
 
 | | Lectin Domain of Lectinolysin complexed with Lewis Y Antigen | | Descriptor: | CALCIUM ION, NICKEL (II) ION, Platelet aggregation factor Sm-hPAF, ... | | Authors: | Feil, S.C. | | Deposit date: | 2010-01-14 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of the lectin regulatory domain of the cholesterol-dependent cytolysin lectinolysin reveals the basis for its lewis antigen specificity.
Structure, 20, 2012
|
|
3LEK
 
 | | Lectin Domain of Lectinolysin complexed with Lewis B Antigen | | Descriptor: | CALCIUM ION, NICKEL (II) ION, Platelet aggregation factor Sm-hPAF, ... | | Authors: | Feil, S.C. | | Deposit date: | 2010-01-15 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the lectin regulatory domain of the cholesterol-dependent cytolysin lectinolysin reveals the basis for its lewis antigen specificity.
Structure, 20, 2012
|
|
4KRM
 
 | |
2YFT
 
 | | Crystal structure of inulosucrase from Lactobacillus johnsonii NCC533 in complex with 1-kestose | | Descriptor: | ACETATE ION, CALCIUM ION, LEVANSUCRASE, ... | | Authors: | Pijning, T, Anwar, M.A, Leemhuis, H, Kralj, S, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2011-04-07 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Inulosucrase from Lactobacillus: Insights Into the Substrate Specificity and Product Specificity of Gh68 Fructansucrases.
J.Mol.Biol., 412, 2011
|
|
3W3N
 
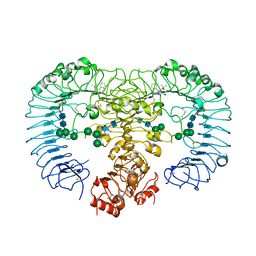 | | Crystal structure of human TLR8 in complex with Resiquimod (R848) crystal form 3 | | Descriptor: | 1-[4-amino-2-(ethoxymethyl)-1H-imidazo[4,5-c]quinolin-1-yl]-2-methylpropan-2-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2012-12-22 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural reorganization of the Toll-like receptor 8 dimer induced by agonistic ligands
Science, 339, 2013
|
|
3W6D
 
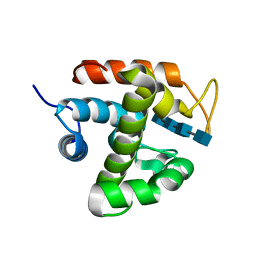 | | Crystal structure of catalytic domain of chitinase from Ralstonia sp. A-471 (E141Q) in complex with tetrasaccharide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysozyme-like chitinolytic enzyme | | Authors: | Arimori, T, Kawamoto, N, Okazaki, N, Nakazawa, M, Miyatake, K, Fukamizo, T, Ueda, M, Tamada, T. | | Deposit date: | 2013-02-14 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures of the Catalytic Domain of a Novel Glycohydrolase Family 23 Chitinase from Ralstonia sp. A-471 Reveals a Unique Arrangement of the Catalytic Residues for Inverting Chitin Hydrolysis
J.Biol.Chem., 288, 2013
|
|
