4P78
 
 | | HicA3 and HicB3 toxin-antitoxin complex | | Descriptor: | GLYCEROL, HicA3 Toxin, HicB3 antitoxin | | Authors: | Li de la Sierra-Gallay, I, Bibi-Triki, S, van Tilbeurgh, H, Lazar, N, Pradel, E. | | Deposit date: | 2014-03-26 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Functional and Structural Analysis of HicA3-HicB3, a Novel Toxin-Antitoxin System of Yersinia pestis.
J.Bacteriol., 196, 2014
|
|
1NJB
 
 | | THYMIDYLATE SYNTHASE | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1996-01-23 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Partitioning roles of side chains in affinity, orientation, and catalysis with structures for mutant complexes: asparagine-229 in thymidylate synthase.
Biochemistry, 35, 1996
|
|
4P7D
 
 | | Antitoxin HicB3 crystal structure | | Descriptor: | Antitoxin HicB3, CHLORIDE ION | | Authors: | Li de la Sierra-Gallay, I, Bibi-Triki, S, van Tilbeurgh, H, Lazar, N, Pradel, E. | | Deposit date: | 2014-03-27 | | Release date: | 2014-08-27 | | Last modified: | 2015-02-04 | | Method: | X-RAY DIFFRACTION (2.781 Å) | | Cite: | Functional and Structural Analysis of HicA3-HicB3, a Novel Toxin-Antitoxin System of Yersinia pestis.
J.Bacteriol., 196, 2014
|
|
1NJC
 
 | | THYMIDYLATE SYNTHASE, MUTATION, N229D WITH 2'-DEOXYCYTIDINE 5'-MONOPHOSPHATE (DCMP) | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1996-01-23 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Partitioning roles of side chains in affinity, orientation, and catalysis with structures for mutant complexes: asparagine-229 in thymidylate synthase.
Biochemistry, 35, 1996
|
|
4PKS
 
 | | Anthrax toxin lethal factor with bound small molecule inhibitor 11 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Lethal factor, ... | | Authors: | Maize, K.M, De la Mora, T, Finzel, B.C. | | Deposit date: | 2014-05-15 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Anthrax toxin lethal factor domain 3 is highly mobile and responsive to ligand binding.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1NCL
 
 | |
4P7F
 
 | | Mouse apo-COMT | | Descriptor: | Catechol O-methyltransferase, HYDROGENPHOSPHATE ION, PHOSPHATE ION, ... | | Authors: | Ehler, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2014-03-27 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Mapping the conformational space accessible to catechol-O-methyltransferase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1MVB
 
 | |
4P7M
 
 | |
1NJA
 
 | | THYMIDYLATE SYNTHASE, MUTATION, N229C WITH 2'-DEOXYCYTIDINE 5'-MONOPHOSPHATE (DCMP) | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1996-01-23 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Partitioning roles of side chains in affinity, orientation, and catalysis with structures for mutant complexes: asparagine-229 in thymidylate synthase.
Biochemistry, 35, 1996
|
|
4PL6
 
 | |
1MUN
 
 | |
4P7U
 
 | |
1MSF
 
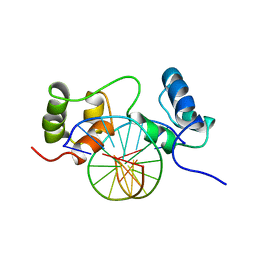 | | SOLUTION STRUCTURE OF A SPECIFIC DNA COMPLEX OF THE MYB DNA-BINDING DOMAIN WITH COOPERATIVE RECOGNITION HELICES | | Descriptor: | C-Myb DNA-Binding Domain, DNA (5'-D(*AP*TP*GP*TP*GP*TP*GP*TP*CP*AP*GP*TP*TP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*AP*AP*CP*TP*GP*AP*CP*AP*CP*AP*CP*AP*T)-3') | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Sekikawa, A, Inoue, T, Kanai, H, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-01-24 | | Release date: | 1995-03-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices.
Cell(Cambridge,Mass.), 79, 1994
|
|
4PLA
 
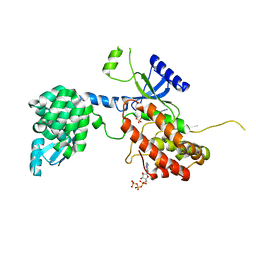 | |
1NJD
 
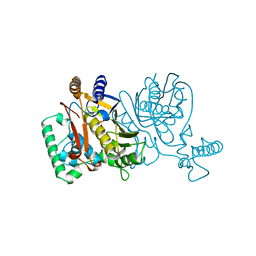 | | THYMIDYLATE SYNTHASE, MUTATION, N229D WITH 2'-DEOXYURIDINE 5'-MONOPHOSPHATE (DUMP) | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1996-01-23 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Partitioning roles of side chains in affinity, orientation, and catalysis with structures for mutant complexes: asparagine-229 in thymidylate synthase.
Biochemistry, 35, 1996
|
|
4P7Y
 
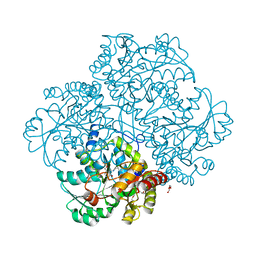 | | L-methionine gamma-lyase from Citrobacter freundii with Y58F substitution | | Descriptor: | DI(HYDROXYETHYL)ETHER, METHIONINE GAMMA-LYASE, PENTAETHYLENE GLYCOL | | Authors: | Revtovich, S.V, Nikulin, A.D, Anufrieva, N.V, Morozova, E.A, Demidkina, T.V. | | Deposit date: | 2014-03-28 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of L-methionine gamma-lyase from Citrobacter freundii with Y58F substitution
To Be Published
|
|
1MVA
 
 | |
4PLH
 
 | | Crystal structure of ancestral apicomplexan malate dehydrogenase with oxamate. | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, OXAMIC ACID, SODIUM ION, ... | | Authors: | Boucher, J.I, Jacobowitz, J.R, Beckett, B.C, Classen, S, Theobald, D.L. | | Deposit date: | 2014-05-16 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An atomic-resolution view of neofunctionalization in the evolution of apicomplexan lactate dehydrogenases.
Elife, 3, 2014
|
|
1MYO
 
 | | SOLUTION STRUCTURE OF MYOTROPHIN, NMR, 44 STRUCTURES | | Descriptor: | MYOTROPHIN | | Authors: | Yang, Y, Nanduri, S, Sen, S, Qin, J. | | Deposit date: | 1998-08-17 | | Release date: | 1999-08-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structural basis of ankyrin-like repeat function as revealed by the solution structure of myotrophin.
Structure, 6, 1998
|
|
4P8E
 
 | | Structure of ribB complexed with substrate (Ru5P) and metal ions | | Descriptor: | 1,2-ETHANEDIOL, 3,4-dihydroxy-2-butanone 4-phosphate synthase, RIBULOSE-5-PHOSPHATE, ... | | Authors: | Islam, Z, Kumar, A, Singh, S, Salmon, L, Karthikeyan, S. | | Deposit date: | 2014-03-31 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis for Competitive Inhibition of 3,4-Dihydroxy-2-butanone-4-phosphate Synthase from Vibrio cholerae.
J.Biol.Chem., 290, 2015
|
|
1NGR
 
 | |
4PLS
 
 | | Crystal Structures of Designed Armadillo Repeat Proteins: Implications of Construct Design and Crystallization Conditions on Overall Structure. | | Descriptor: | ACETATE ION, Arm00010, CALCIUM ION | | Authors: | Reichen, C, Madhurantakam, C, Plueckthun, A, Mittl, P.R. | | Deposit date: | 2014-05-19 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structures of Designed Armadillo Repeat Proteins: Implications of Construct Design and Crystallization Conditions on Overall Structure.
Protein Sci., 23, 2014
|
|
1NAO
 
 | | SOLUTION STRUCTURE OF AN RNA 2'-O-METHYLATED RNA DUPLEX CONTAINING AN RNA/DNA HYBRID SEGMENT AT THE CENTER, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA/RNA (5'-R(*OMGP*OMUP*OMC)-D(P*AP*TP*CP*T)-R(P*OMCP*OMC)-3'), RNA (5'-R(*GP*GP*AP*GP*AP*UP*GP*AP*C)-3') | | Authors: | Nishizaki, T, Iwai, S, Ohtsuka, E, Nakamura, H. | | Deposit date: | 1996-03-29 | | Release date: | 1997-01-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an RNA.2'-O-methylated RNA hybrid duplex containing an RNA.DNA hybrid segment at the center.
Biochemistry, 36, 1997
|
|
4P8N
 
 | | Crystal structure of M. tuberculosis DprE1 in complex with the non-covalent inhibitor QN118 | | Descriptor: | 3-[(3-fluoro-4-methoxybenzyl)amino]-6-(trifluoromethyl)quinoxaline-2-carboxylic acid, FLAVIN-ADENINE DINUCLEOTIDE, IMIDAZOLE, ... | | Authors: | Neres, J, Pojer, F, Cole, S.T. | | Deposit date: | 2014-03-31 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | 2-Carboxyquinoxalines Kill Mycobacterium tuberculosis through Noncovalent Inhibition of DprE1.
Acs Chem.Biol., 10, 2015
|
|
