4BKK
 
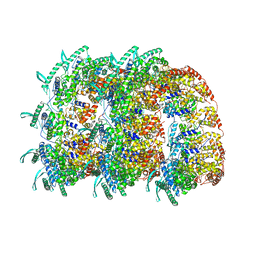 | | The Respiratory Syncytial Virus nucleoprotein-RNA complex forms a left-handed helical nucleocapsid. | | Descriptor: | NUCLEOPROTEIN, RNA (161-MER) | | Authors: | Bakker, S.E, Duquerroy, S, Galloux, M, Loney, C, Conner, E, Eleouet, J.F, Rey, F.A, Bhella, D. | | Deposit date: | 2013-04-26 | | Release date: | 2013-10-30 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY | | Cite: | The Respiratory Syncytial Virus Nucleoprotein-RNA Complex Forms a Left-Handed Helical Nucleocapsid.
J.Gen.Virol., 94, 2013
|
|
2OHH
 
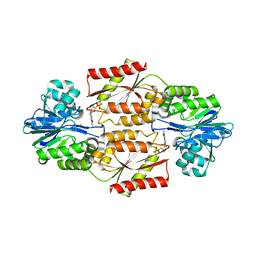 | | Crystal Structure of coenzyme F420H2 oxidase (FprA), a diiron flavoprotein, active oxidized state | | Descriptor: | FE (III) ION, FLAVIN MONONUCLEOTIDE, SULFATE ION, ... | | Authors: | Seedorf, H, Warkentin, E, Ermler, U. | | Deposit date: | 2007-01-10 | | Release date: | 2007-05-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of coenzyme F420H2 oxidase (FprA), a di-iron flavoprotein from methanogenic Archaea catalyzing the reduction of O2 to H2O.
Febs J., 274, 2007
|
|
7YHJ
 
 | |
6SD2
 
 | | Structure of the RBM2inner region of the Salmonella flagella MS-ring protein FliF with 21-fold symmetry applied. | | Descriptor: | Flagellar M-ring protein | | Authors: | Johnson, S, Fong, Y.H, Deme, J.C, Furlong, E.J, Kuhlen, L, Lea, S.M. | | Deposit date: | 2019-07-26 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Symmetry mismatch in the MS-ring of the bacterial flagellar rotor explains the structural coordination of secretion and rotation.
Nat Microbiol, 5, 2020
|
|
4FQO
 
 | | Crystal Structure of Calcium-Loaded S100B Bound to SBi4211 | | Descriptor: | 4,4'-[heptane-1,7-diylbis(oxy)]dibenzenecarboximidamide, CALCIUM ION, Protein S100-B | | Authors: | McKnight, L.E, Raman, E.P, Bezawada, P, Kudrimoti, S, Wilder, P.T, Hartman, K.G, Toth, E.A, Coop, A, MacKerrell, A.D, Weber, D.J. | | Deposit date: | 2012-06-25 | | Release date: | 2012-10-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Based Discovery of a Novel Pentamidine-Related Inhibitor of the Calcium-Binding Protein S100B.
ACS Med Chem Lett, 3, 2012
|
|
1DLJ
 
 | | THE FIRST STRUCTURE OF UDP-GLUCOSE DEHYDROGENASE (UDPGDH) REVEALS THE CATALYTIC RESIDUES NECESSARY FOR THE TWO-FOLD OXIDATION | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Campbell, R.E, Mosimann, S.C, van de Rijn, I, Tanner, M.E, Strynadka, N.C.J. | | Deposit date: | 1999-12-09 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The first structure of UDP-glucose dehydrogenase reveals the catalytic residues necessary for the two-fold oxidation.
Biochemistry, 39, 2000
|
|
6BUZ
 
 | | Cryo-EM structure of CENP-A nucleosome in complex with kinetochore protein CENP-N | | Descriptor: | DNA (147-MER), Histone H2A, Histone H2B, ... | | Authors: | Chittori, S, Hong, J, Kelly, A.E, Bai, Y, Subramaniam, S. | | Deposit date: | 2017-12-11 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structural mechanisms of centromeric nucleosome recognition by the kinetochore protein CENP-N.
Science, 359, 2018
|
|
6SBS
 
 | | YtrA from Sulfolobus acidocaldarius, a GntR-family transcription factor | | Descriptor: | Regulatory protein | | Authors: | Lemmens, L, Valegard, K, Lindas, A.C, Peeters, E, Maes, D. | | Deposit date: | 2019-07-22 | | Release date: | 2019-07-31 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | YtrASa, a GntR-Family Transcription Factor, Represses Two Genetic Loci Encoding Membrane Proteins inSulfolobus acidocaldarius.
Front Microbiol, 10, 2019
|
|
1OOP
 
 | | The Crystal Structure of Swine Vesicular Disease Virus | | Descriptor: | Coat protein VP1, Coat protein VP2, Coat protein VP3, ... | | Authors: | Fry, E.E, Knowles, N.J, Newman, J.W.I, Wilsden, G, Rao, Z, King, A.M.Q, Stuart, D.I. | | Deposit date: | 2003-03-04 | | Release date: | 2003-04-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Swine Vesicular Disease Virus and Implications for Host Adaptation
J.Virol., 77, 2003
|
|
7JJC
 
 | |
7Y0G
 
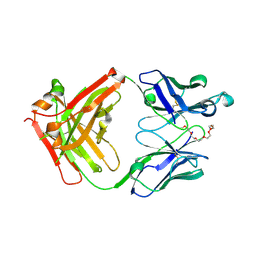 | | Crystal structure of anti-mPEG h15-2b Fab | | Descriptor: | 15-2b heavy chain, 15-2b light chain, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL | | Authors: | Chang, C.Y, Nguyen, T.M.T, Lin, E.C, Su, Y.C. | | Deposit date: | 2022-06-05 | | Release date: | 2022-08-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural determination of an antibody that specifically recognizes polyethylene glycol with a terminal methoxy group.
Commun Chem, 5, 2022
|
|
7JK8
 
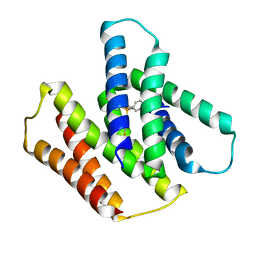 | | EmrE S64V mutant bound to tetra(4-fluorophenyl)phosphonium at pH 5.8 | | Descriptor: | Multidrug SMR transporter, tetrakis(4-fluorophenyl)phosphanium | | Authors: | Shcherbakov, A.A, Hisao, G, Mandala, V.S, Thomas, N.E, Soltani, M, Salter, E.A, Davis Jr, J.H, Henzler-Wildman, K.A, Hong, M. | | Deposit date: | 2020-07-27 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Structure and dynamics of the drug-bound bacterial transporter EmrE in lipid bilayers.
Nat Commun, 12, 2021
|
|
7XW6
 
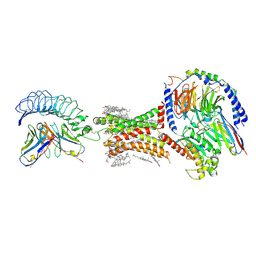 | | TSHR-Gs-M22 antibody-ML109 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Duan, J, Xu, P, Luan, X, Ji, Y, Yuan, Q, He, X, Ye, J, Cheng, X, Jiang, H, Zhang, S, Jiang, Y, Xu, H.E. | | Deposit date: | 2022-05-26 | | Release date: | 2022-08-17 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Hormone- and antibody-mediated activation of the thyrotropin receptor.
Nature, 609, 2022
|
|
4ZC9
 
 | | Crystal Structure of the BRD4a/DB-2-190 complex | | Descriptor: | 2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]-N-(4-{[({2-[(3S)-2,6-dioxopiperidin-3-yl]-1,3-dioxo-2,3-dihydro-1H-isoindol-4-yl}oxy)acetyl]amino}butyl)acetamide, Bromodomain-containing protein 4 | | Authors: | Seo, H.-S, DeAngelo, S, Bradner, J.E. | | Deposit date: | 2015-04-15 | | Release date: | 2015-11-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | DRUG DEVELOPMENT. Phthalimide conjugation as a strategy for in vivo target protein degradation.
Science, 348, 2015
|
|
6TN4
 
 | | Rapid optimisation of fragments and hits to lead compounds from screening of crude reaction mixtures | | Descriptor: | 2,4-bis(oxidanyl)benzamide, CHLORIDE ION, Heat shock protein HSP 90-alpha | | Authors: | Baker, L.M, Aimon, A, Murray, J.B, Surgenor, A.E, Matassova, N, Roughley, S.D, von Delft, F, Hubbard, R.E. | | Deposit date: | 2019-12-05 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.274 Å) | | Cite: | Rapid optimisation of fragments and hits to lead compounds from screening of crude reaction mixtures
Commun Chem, 2020
|
|
2HQS
 
 | | Crystal structure of TolB/Pal complex | | Descriptor: | ACETATE ION, GLYCEROL, Peptidoglycan-associated lipoprotein, ... | | Authors: | Grishkovskaya, I, Bonsor, D.A, Kleanthous, C, Dodson, E.J. | | Deposit date: | 2006-07-19 | | Release date: | 2007-04-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular mimicry enables competitive recruitment by a natively disordered protein.
J.Am.Chem.Soc., 129, 2007
|
|
6SHN
 
 | | Escherichia coli AGPase in complex with FBP. Symmetry C1 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Glucose-1-phosphate adenylyltransferase | | Authors: | Cifuente, J.O, Comino, N, D'Angelo, C, Marina, A, Gil-Carton, D, Albesa-Jove, D, Guerin, M.E. | | Deposit date: | 2019-08-07 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The allosteric control mechanism of bacterial glycogen biosynthesis disclosed by cryoEM.
Curr Res Struct Biol, 2, 2020
|
|
6SID
 
 | |
4ZGK
 
 | | Structure of Mdm2 with low molecular weight inhibitor. | | Descriptor: | (5R)-3,5-bis(4-chlorobenzyl)-4-(6-chloro-1H-indol-3-yl)-5-hydroxyfuran-2(5H)-one, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Twarda-Clapa, A, Zak, K.M, Wrona, E.M, Grudnik, P, Dubin, G, Holak, T.A. | | Deposit date: | 2015-04-23 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Unique Mdm2-Binding Mode of the 3-Pyrrolin-2-one- and 2-Furanone-Based Antagonists of the p53-Mdm2 Interaction.
ACS Chem. Biol., 11, 2016
|
|
226L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | Deposit date: | 1997-06-25 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
1MBO
 
 | |
7JG0
 
 | | Human GAR transformylase in complex with GAR substrate and AGF102 inhibitor | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, N-{5-[4-(2-amino-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidin-6-yl)butyl]thiophene-2-carbonyl}-L-glutamic acid, Trifunctional purine biosynthetic protein adenosine-3 | | Authors: | Wong-Roushar, J, Dann III, C.E. | | Deposit date: | 2020-07-18 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Discovery of 6-substituted thieno[2,3-d]pyrimidine analogs as dual inhibitors of glycinamide ribonucleotide formyltransferase and 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase in de novo purine nucleotide biosynthesis in folate receptor expressing human tumors
Bioorg.Med.Chem., 37, 2021
|
|
1USN
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN FIBROBLAST STROMELYSIN-1 INHIBITED WITH THIADIAZOLE INHIBITOR PNU-142372 | | Descriptor: | 2-[3-(5-MERCAPTO-[1,3,4]THIADIAZOL-2YL)-UREIDO]-N-METHYL-3-PENTAFLUOROPHENYL-PROPIONAMIDE, CALCIUM ION, STROMELYSIN-1, ... | | Authors: | Finzel, B.C, Bryant Junior, G.L, Baldwin, E.T. | | Deposit date: | 1998-06-09 | | Release date: | 1998-12-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterizations of nonpeptidic thiadiazole inhibitors of matrix metalloproteinases reveal the basis for stromelysin selectivity.
Protein Sci., 7, 1998
|
|
7YKE
 
 | | Crystal structure of chondroitin ABC lyase I in complex with chondroitin disaccharide 4,6-sulfate | | Descriptor: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4,6-di-O-sulfo-beta-D-galactopyranose, Chondroitin sulfate ABC endolyase, MAGNESIUM ION | | Authors: | Takashima, M, Watanabe, I, Miyanaga, A, Eguchi, T. | | Deposit date: | 2022-07-22 | | Release date: | 2022-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Biochemical and crystallographic assessments of the effect of 4,6-O-disulfated disaccharide moieties in chondroitin sulfate E on chondroitinase ABC I activity.
Febs J., 290, 2023
|
|
7MXY
 
 | | Cryo-EM structure of PfFNT-inhibitor complex | | Descriptor: | (Z)-4,4,5,5,5-pentakis(fluoranyl)-1-(4-methoxy-2-oxidanyl-phenyl)-3-oxidanyl-pent-2-en-1-one, Formate-nitrite transporter | | Authors: | Yu, E.W, Su, C, Lyu, M. | | Deposit date: | 2021-05-19 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.18 Å) | | Cite: | Structural basis of transport and inhibition of the Plasmodium falciparum transporter PfFNT
EMBO Rep, 22, 2021
|
|
