5TY9
 
 | | Identification of a New Zinc Binding Chemotype by Fragment Screening | | Descriptor: | (5R)-5-(2,4-dimethoxyphenyl)-1,3-oxazolidine-2,4-dione, Carbonic anhydrase 2, ZINC ION | | Authors: | Peat, T.S, Poulsen, S.A, Ren, B, Dolezal, O, Woods, L.A, Mujumdar, P, Chrysanthopoulos, P.K. | | Deposit date: | 2016-11-18 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Identification of a New Zinc Binding Chemotype by Fragment Screening.
J. Med. Chem., 60, 2017
|
|
6WSB
 
 | |
1QA5
 
 | |
7CCS
 
 | | Consensus mutated xCT-CD98hc complex | | Descriptor: | 4F2 cell-surface antigen heavy chain, Consensus mutated Anionic Amino Acid Transporter Light Chain, Xc- System | | Authors: | Oda, K, Lee, Y, Takemoto, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2020-06-17 | | Release date: | 2020-12-09 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Consensus mutagenesis approach improves the thermal stability of system x c - transporter, xCT, and enables cryo-EM analyses.
Protein Sci., 29, 2020
|
|
7C79
 
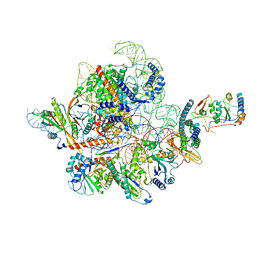 | | Cryo-EM structure of yeast Ribonuclease MRP | | Descriptor: | MAGNESIUM ION, RNases MRP/P 32.9 kDa subunit, Ribonuclease MRP RNA subunit NME1, ... | | Authors: | Lan, P, Wu, J, Lei, M. | | Deposit date: | 2020-05-24 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural insight into precursor ribosomal RNA processing by ribonuclease MRP.
Science, 369, 2020
|
|
7CM4
 
 | | Crystal Structure of COVID-19 virus spike receptor-binding domain complexed with a neutralizing antibody CT-P59 | | Descriptor: | 1,2-ETHANEDIOL, IgG heavy chain, IgG light chain, ... | | Authors: | Kim, Y.G, Jeong, J.H, Bae, J.S, Lee, J. | | Deposit date: | 2020-07-24 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein.
Nat Commun, 12, 2021
|
|
7CPM
 
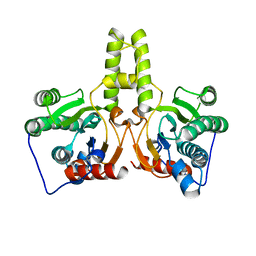 | | CRYSTAL STRUCTURE OF DODECAPRENYL DIPHOSPHATE SYNTHASE FROM THERMOBIFIDA FUSCA | | Descriptor: | Trans,polycis-polyprenyl diphosphate synthase ((2Z,6E)-farnesyl diphosphate specific) | | Authors: | Kurokawa, H, Ambo, T, Takahashi, S, Koyama, T. | | Deposit date: | 2020-08-07 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Thermobifida fusca cis-prenyltransferase reveals the dynamic nature of its RXG motif-mediated inter-subunit interactions critical for its catalytic activity.
Biochem.Biophys.Res.Commun., 532, 2020
|
|
7CPN
 
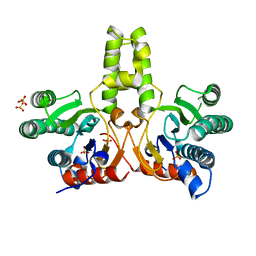 | | CRYSTAL STRUCTURE OF DODECAPRENYL DIPHOSPHATE SYNTHASE FROM THERMOBIFIDA FUSCA | | Descriptor: | GLYCEROL, SULFATE ION, Trans,polycis-polyprenyl diphosphate synthase ((2Z,6E)-farnesyl diphosphate specific) | | Authors: | Kurokawa, H, Ambo, T, Takahasi, S, Koyama, T. | | Deposit date: | 2020-08-07 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal structure of Thermobifida fusca cis-prenyltransferase reveals the dynamic nature of its RXG motif-mediated inter-subunit interactions critical for its catalytic activity.
Biochem.Biophys.Res.Commun., 532, 2020
|
|
6WKA
 
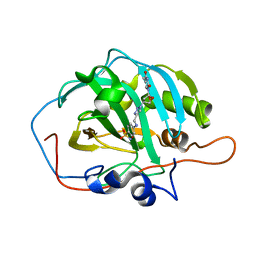 | | human carbonic anhydrase II bound to an inhibitor modified with azidothymidine | | Descriptor: | 3'-deoxy-3'-(4-{[(4-sulfamoylphenyl)amino]methyl}-1H-1,2,3-triazol-1-yl)thymidine, Carbonic anhydrase 2, ZINC ION | | Authors: | Peat, T.S. | | Deposit date: | 2020-04-15 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Azidothymidine "Clicked" into 1,2,3-Triazoles: First Report on Carbonic Anhydrase-Telomerase Dual-Hybrid Inhibitors.
J.Med.Chem., 63, 2020
|
|
8E3Z
 
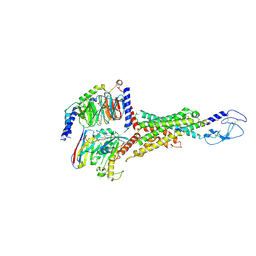 | | Cryo-EM structure of the VPAC1R-VIP-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Piper, S.J, Danev, R, Sexton, P, Wootten, D. | | Deposit date: | 2022-08-17 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Understanding VPAC receptor family peptide binding and selectivity.
Nat Commun, 13, 2022
|
|
7UIN
 
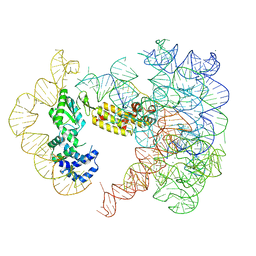 | | CryoEM Structure of an Group II Intron Retroelement | | Descriptor: | AMMONIUM ION, DNA (37-MER), E.r IIC Intron, ... | | Authors: | Chung, K, Xu, L. | | Deposit date: | 2022-03-29 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of a mobile intron retroelement poised to attack its structured DNA target
Science, 378, 2022
|
|
1LCX
 
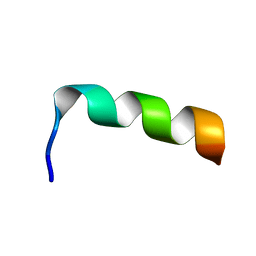 | | NMR structure of HIV-1 gp41 659-671 13mer peptide | | Descriptor: | GP41 | | Authors: | Biron, Z, Khare, S, Samson, A.O, Hayek, Y, Naider, F, Anglister, J. | | Deposit date: | 2002-04-07 | | Release date: | 2002-12-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Monomeric 3(10)-Helix Is Formed in Water by a 13-Residue Peptide
Representing the Neutralizing Determinant of HIV-1 on gp41.
Biochemistry, 41, 2002
|
|
8U9R
 
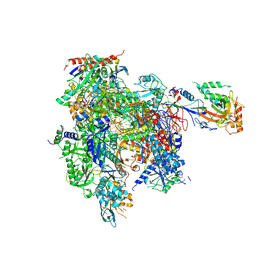 | | STRUCTURAL BASIS OF TRANSCRIPTION: RNA POLYMERASE II SUBSTRATE BINDING AND METAL COORDINATION USING A FREE-ELECTRON LASER | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*CP*AP*CP*GP*TP*CP*CP*CP*TP*CP*TP*CP*GP*A)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Arjunan, P, Calero, G, Kaplan, C.D. | | Deposit date: | 2023-09-20 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Structural basis of transcription: RNA polymerase II substrate binding and metal coordination using a free-electron laser.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7CM3
 
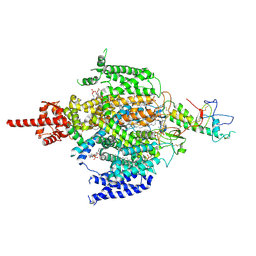 | | Cryo-EM structure of human NALCN in complex with FAM155A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, J, Yan, Z, Ke, M. | | Deposit date: | 2020-07-24 | | Release date: | 2020-11-11 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the human sodium leak channel NALCN in complex with FAM155A.
Nat Commun, 11, 2020
|
|
7UT5
 
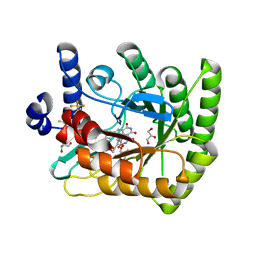 | | Acinetobacter baumannii dihydroorotate dehydrogenase bound with inhibitor DSM186 | | Descriptor: | (4R)-7-methyl-N-[4-(pentafluoro-lambda~6~-sulfanyl)phenyl]imidazo[1,2-a]pyrimidin-5-amine, Dihydroorotate dehydrogenase (quinone), FLAVIN MONONUCLEOTIDE, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2022-04-26 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Repurposed dihydroorotate dehydrogenase inhibitors with efficacy against drug-resistant Acinetobacter baumannii.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6X28
 
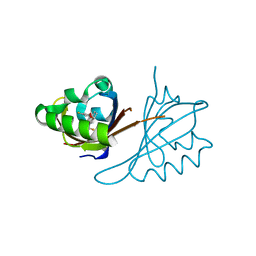 | | Crystal structure of PT2243 bound to HIF2a-B*:ARNT-B* complex | | Descriptor: | (1R)-4-(3,5-difluorophenoxy)-7-(trifluoromethyl)-2,3-dihydro-1H-inden-1-ol, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Du, X. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of PT2243 bound to HIF2-PasB*:ARNT-PasB* complex
To Be Published
|
|
6X37
 
 | | Crystal structure of PT3245 bound to HIF2a-B*:ARNT-B* complex | | Descriptor: | 3-fluoro-5-{[(7R)-7-hydroxy-1-(trifluoromethyl)-6,7-dihydro-5H-cyclopenta[c]pyridin-4-yl]oxy}benzonitrile, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Du, X. | | Deposit date: | 2020-05-21 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of PT3245 bound to HIF2a-B*:ARNT-B* complex
To Be Published
|
|
7UCD
 
 | | Transcription factor FosB/JunD bZIP domain covalently modified with the cysteine-targeting alpha-haloketone compound Z2159931480 | | Descriptor: | 7-acetyl-4-methoxy-1-benzofuran-3(2H)-one, CHLORIDE ION, Protein fosB, ... | | Authors: | Kumar, A, Machius, M.C, Rudenko, G. | | Deposit date: | 2022-03-16 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Chemically targeting the redox switch in AP1 transcription factor Delta FOSB.
Nucleic Acids Res., 50, 2022
|
|
6X21
 
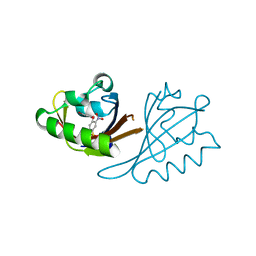 | | Crystal structure of PT1673 bound to HIF2a-B*:ARNT-B* complex | | Descriptor: | 1-(3-bromo-5-fluorophenoxy)-4-[(difluoromethyl)sulfonyl]-2-nitrobenzene, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Du, X. | | Deposit date: | 2020-05-19 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of PT1673 bound to HIF2a-B*:ARNT-B* complex
To Be Published
|
|
6X3D
 
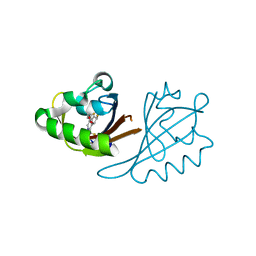 | | Crystal structure of PT3388 bound to HIF2a-B*:ARNT-B* complex | | Descriptor: | (6R,7S)-4-[(3,3-difluorocyclobutyl)oxy]-6-fluoro-1-(trifluoromethyl)-6,7-dihydro-5H-cyclopenta[c]pyridin-7-ol, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Du, X. | | Deposit date: | 2020-05-21 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of PT3388 bound to HIF2a-B*:ARNT-B* complex
To Be Published
|
|
8E86
 
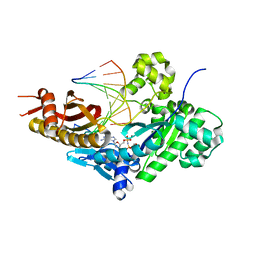 | | Human DNA polymerase eta-DNA-rC-ended primer-dGMPNPP ternary mismatch complex with Mn2+ | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*CP*AP*TP*TP*GP*TP*GP*AP*CP*GP*CP*T)-3'), DNA polymerase eta, ... | | Authors: | Chang, C, Gao, Y. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Primer terminal ribonucleotide alters the active site dynamics of DNA polymerase eta and reduces DNA synthesis fidelity.
J.Biol.Chem., 299, 2023
|
|
8E8A
 
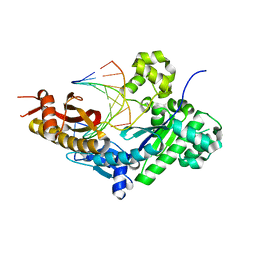 | | Human DNA polymerase eta-DNA-dT-ended primer binary complex | | Descriptor: | DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*TP*AP*TP*GP*AP*CP*GP*CP*T)-3'), DNA polymerase eta | | Authors: | Chang, C, Gao, Y. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Primer terminal ribonucleotide alters the active site dynamics of DNA polymerase eta and reduces DNA synthesis fidelity.
J.Biol.Chem., 299, 2023
|
|
8E87
 
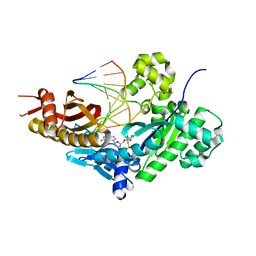 | | Human DNA polymerase eta-DNA-rA-ended primer-dGMPNPP ternary mismatch complex with Mg2+ | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*CP*AP*TP*TP*TP*TP*GP*AP*CP*GP*CP*T)-3'), DNA polymerase eta, ... | | Authors: | Chang, C, Gao, Y. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Primer terminal ribonucleotide alters the active site dynamics of DNA polymerase eta and reduces DNA synthesis fidelity.
J.Biol.Chem., 299, 2023
|
|
8E8J
 
 | | Human DNA polymerase eta-DNA-rG-ended primer-dGMPNPP ternary mismatch complex with Mg2+ | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]guanosine, DNA (5'-D(*CP*AP*TP*TP*CP*TP*GP*AP*CP*GP*CP*T)-3'), DNA polymerase eta, ... | | Authors: | Chang, C, Gao, Y. | | Deposit date: | 2022-08-25 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Primer terminal ribonucleotide alters the active site dynamics of DNA polymerase eta and reduces DNA synthesis fidelity.
J.Biol.Chem., 299, 2023
|
|
8E8B
 
 | |
