8VJ7
 
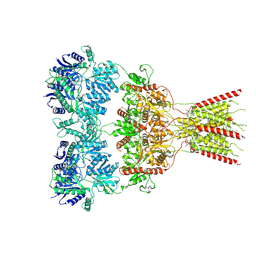 | | GluA2 bound to GYKI-52466 and Glutamate, Inhibited State 2 | | Descriptor: | 4-[(5S,8R)-8-methyl-6,7,8,9-tetrahydro-2H,5H-[1,3]dioxolo[4,5-h][2,3]benzodiazepin-5-yl]aniline, GLUTAMIC ACID, Isoform Flip of Glutamate receptor 2 | | Authors: | Hale, W.D, Montano Romero, A, Huganir, R.L, Twomey, E.C. | | Deposit date: | 2024-01-05 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.85 Å) | | Cite: | Allosteric competition and inhibition in AMPA receptors.
Nat.Struct.Mol.Biol., 2024
|
|
5VCV
 
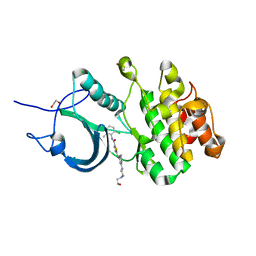 | | CRYSTAL STRUCTURE OF HUMAN MYT1 KINASE DOMAIN IN COMPLEX WITH Dasatinib | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase, ... | | Authors: | Zhu, J.-Y, Martin, M.P, Schonbrunn, E. | | Deposit date: | 2017-04-01 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Basis of Wee Kinases Functionality and Inactivation by Diverse Small Molecule Inhibitors.
J. Med. Chem., 60, 2017
|
|
5IOV
 
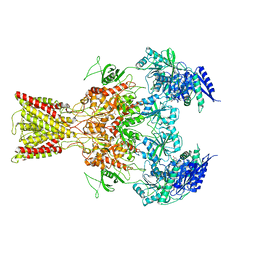 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the glutamate/glycine/Ro25-6981-bound conformation | | Descriptor: | 4-[(1R,2S)-3-(4-benzylpiperidin-1-yl)-1-hydroxy-2-methylpropyl]phenol, GLUTAMIC ACID, GLYCINE, ... | | Authors: | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, Mchaourab, S.H, Gouaux, E. | | Deposit date: | 2016-03-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
4N7I
 
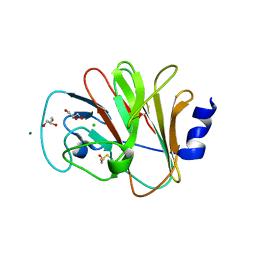 | | Crystal Structure of Intracellular B30.2 Domain of BTN3A1 | | Descriptor: | BETA-MERCAPTOETHANOL, Butyrophilin subfamily 3 member A1, CHLORIDE ION, ... | | Authors: | Sandstrom, A, Adams, E.J. | | Deposit date: | 2013-10-15 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4015 Å) | | Cite: | The Intracellular B30.2 Domain of Butyrophilin 3A1 Binds Phosphoantigens to Mediate Activation of Human V gamma 9V delta 2 T Cells.
Immunity, 40, 2014
|
|
5VDK
 
 | | Crystal structure of human WEE2 kinase domain in complex with MK1775 | | Descriptor: | 1-[6-(2-hydroxypropan-2-yl)pyridin-2-yl]-6-{[4-(4-methylpiperazin-1-yl)phenyl]amino}-2-(prop-2-en-1-yl)-1,2-dihydro-3H-pyrazolo[3,4-d]pyrimidin-3-one, PHOSPHATE ION, Wee1-like protein kinase 2 | | Authors: | Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2017-04-03 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis of Wee Kinases Functionality and Inactivation by Diverse Small Molecule Inhibitors.
J. Med. Chem., 60, 2017
|
|
4BOL
 
 | | Crystal structure of AmpDh2 from Pseudomonas aeruginosa in complex with pentapeptide | | Descriptor: | AMPDH2, D-alanyl-N-[(2S,6R)-6-amino-6-carboxy-1-{[(1R)-1-carboxyethyl]amino}-1-oxohexan-2-yl]-D-glutamine, ZINC ION | | Authors: | Artola-Recolons, C, Martinez-Caballero, S, Lee, M, Carrasco-Lopez, C, Hesek, D, Spink, E.E, Lastochkin, E, Zhang, W, Hellman, L.M, Boggess, B, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2013-05-21 | | Release date: | 2013-07-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Reaction Products and the X-Ray Structure of Ampdh2, a Virulence Determinant of Pseudomonas Aeruginosa.
J.Am.Chem.Soc., 135, 2013
|
|
6GB2
 
 | | Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM. This file contains the 39S ribosomal subunit. | | Descriptor: | 'Mitochondrial ribosomal protein L30, 'Mitochondrial ribosomal protein L55, 'Mitochondrial ribosomal protein L59, ... | | Authors: | Kummer, E, Leibundgut, M, Boehringer, D, Ban, N. | | Deposit date: | 2018-04-13 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM.
Nature, 560, 2018
|
|
2C8E
 
 | | Structure of the ARTT motif E214N mutant C3bot1 Exoenzyme (Free state, crystal form III) | | Descriptor: | MONO-ADP-RIBOSYLTRANSFERASE C3, SULFATE ION | | Authors: | Stura, E.A, Menetrey, J, Flatau, G, Boquet, P, Menez, A. | | Deposit date: | 2005-12-03 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for the Nad-Hydrolysis Mechanism and the Artt-Loop Plasticity of C3 Exoenzymes.
Protein Sci., 17, 2008
|
|
6GCW
 
 | | Focal Adhesion Kinase catalytic domain in complex with irreversible inhibitor | | Descriptor: | 2-[[5-chloranyl-2-[[4-[[[1-[2-(propanoylamino)ethyl]-1,2,3-triazol-4-yl]methylamino]methyl]phenyl]amino]pyrimidin-4-yl]amino]-~{N}-methyl-benzamide, Focal adhesion kinase 1, SULFATE ION | | Authors: | Yen-Pon, E, Li, B, Acebron-Garcia-de-Eulate, M, Tomkiewicz-Raulet, C, Dawson, J, Lietha, D, Frame, M.C, Coumoul, X, Garbay, C, Etheve-Quelquejeu, M, Chen, H. | | Deposit date: | 2018-04-19 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design, Synthesis, and Characterization of the First Irreversible Inhibitor of Focal Adhesion Kinase.
Acs Chem.Biol., 13, 2018
|
|
1M6C
 
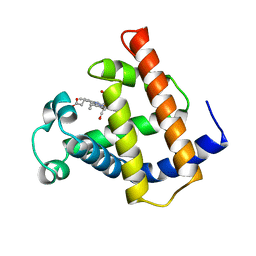 | | V68N MYOGLOBIN WITH CO | | Descriptor: | CARBON MONOXIDE, PROTEIN (MYOGLOBIN), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Murshudov, G.N, Krzywda, S, Brzozowski, A.M, Jaskolski, M, Scott, E.E, Klizas, S.A, Gibson, Q.H, Olson, J.S, Wilkinson, A.J. | | Deposit date: | 1998-08-12 | | Release date: | 1998-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Stabilizing bound O2 in myoglobin by valine68 (E11) to asparagine substitution.
Biochemistry, 37, 1998
|
|
1VTX
 
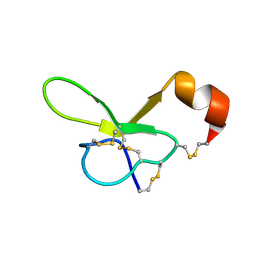 | |
1NOI
 
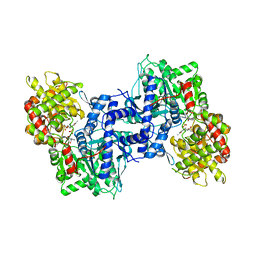 | |
1MKD
 
 | | crystal structure of PDE4D catalytic domain and zardaverine complex | | Descriptor: | 6-(4-DIFLUOROMETHOXY-3-METHOXY-PHENYL)-2H-PYRIDAZIN-3-ONE, MAGNESIUM ION, Phosphodiesterase 4D, ... | | Authors: | Lee, M.E, Markowitz, J, Lee, J.-O, Lee, H. | | Deposit date: | 2002-08-29 | | Release date: | 2003-03-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of phosphodiesterase 4D and inhibitor complex
FEBS LETT., 530, 2002
|
|
7V5L
 
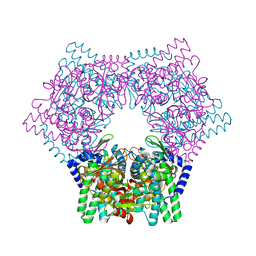 | | Crystal structure of human bleomycin hydrolase | | Descriptor: | Bleomycin hydrolase | | Authors: | Chang, C.Y, Zheng, Y.Z, Huang, S.J, Wang, Y.L, Toh, S.I, Lin, E.C. | | Deposit date: | 2021-08-17 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The Structure-Function Relationship of Human Bleomycin Hydrolase: Mutation of a Cysteine Protease into a Serine Protease.
Chembiochem, 23, 2022
|
|
7MNY
 
 | | Crystal Structure of Nup358/RanBP2 Ran-binding domain 3 in complex with Ran-GPPNHP | | Descriptor: | E3 SUMO-protein ligase RanBP2, GTP-binding nuclear protein Ran, MAGNESIUM ION, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
4CEV
 
 | | ARGINASE FROM BACILLUS CALDEVELOX, L-ORNITHINE COMPLEX | | Descriptor: | GUANIDINE, L-ornithine, MANGANESE (II) ION, ... | | Authors: | Bewley, M.C, Jeffrey, P.D, Patchett, M.L, Kanyo, Z.F, Baker, E.N. | | Deposit date: | 1999-03-15 | | Release date: | 1999-04-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of Bacillus caldovelox arginase in complex with substrate and inhibitors reveal new insights into activation, inhibition and catalysis in the arginase superfamily.
Structure Fold.Des., 7, 1999
|
|
4WVU
 
 | | CRYSTAL STRUCTURE OF XIAP-BIR2 DOMAIN COMPLEXED WITH LIGAND BOUND | | Descriptor: | 3,11-DIFLUORO-6,8,13-TRIMETHYL-8H-QUINO[4,3,2-KL]ACRIDIN-13-IUM, E3 ubiquitin-protein ligase XIAP, GLYCEROL, ... | | Authors: | Pokross, M.E. | | Deposit date: | 2014-11-07 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The Discovery of Macrocyclic XIAP Antagonists from a DNA-Programmed Chemistry Library, and Their Optimization To Give Lead Compounds with in Vivo Antitumor Activity.
J.Med.Chem., 58, 2015
|
|
1W1X
 
 | | Structure of Neuraminidase from English duck subtype N6 complexed with 30 mM sialic acid (NANA, Neu5Ac), crystal soaked for 3 hours at 277 K. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Rudino-Pinera, E, Tunnah, P, Crennell, S.J, Webster, R.G, Laver, W.G, Garman, E.F. | | Deposit date: | 2004-06-24 | | Release date: | 2006-01-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Type a Influenza Virus Neuraminidase of the N6 Subtype Reveals the Existence of Two Separate Neu5Ac Binding Sites
To be Published
|
|
5LDA
 
 | | Structure of deubiquitinating enzyme homolog (Pyrococcus furiosus JAMM1) in complex with ubiquitin-like SAMP2. | | Descriptor: | GLYCEROL, JAMM1, SAMP2, ... | | Authors: | Cao, S, Engilberge, S, Girard, E, Gabel, F, Franzetti, B, Maupin-Furlow, J.A. | | Deposit date: | 2016-06-24 | | Release date: | 2017-06-21 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insight into Ubiquitin-Like Protein Recognition and Oligomeric States of JAMM/MPN(+) Proteases.
Structure, 25, 2017
|
|
8VJ6
 
 | | GluA2 bound to GYKI-52466 and Glutamate, Inhibited State 1 | | Descriptor: | 4-[(5S,8R)-8-methyl-6,7,8,9-tetrahydro-2H,5H-[1,3]dioxolo[4,5-h][2,3]benzodiazepin-5-yl]aniline, Isoform Flip of Glutamate receptor 2 | | Authors: | Hale, W.D, Montano Romero, A, Huganir, R.L, Twomey, E.C. | | Deposit date: | 2024-01-05 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Allosteric competition and inhibition in AMPA receptors.
Nat.Struct.Mol.Biol., 2024
|
|
6BQG
 
 | | Crystal structure of 5-HT2C in complex with ergotamine | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2C,Soluble cytochrome b562, Ergotamine | | Authors: | Peng, Y, McCorvy, J.D, Harpsoe, K, Lansu, K, Yuan, S, Popov, P, Qu, L, Pu, M, Che, T, Nikolajse, L.F, Huang, X.P, Wu, Y, Shen, L, Bjorn-Yoshimoto, W.E, Ding, K, Wacker, D, Han, G.W, Cheng, J, Katritch, V, Jensen, A.A, Hanson, M.A, Zhao, S, Gloriam, D.E, Roth, B.L, Stevens, R.C, Liu, Z. | | Deposit date: | 2017-11-27 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 5-HT2C Receptor Structures Reveal the Structural Basis of GPCR Polypharmacology.
Cell, 172, 2018
|
|
2PDA
 
 | |
5KYM
 
 | | Crystal Structure of the 1-acyl-sn-glycerophosphate (LPA) acyltransferase, PlsC, from Thermotoga maritima | | Descriptor: | 1-HEPTADECANOYL-2-TRIDECANOYL-3-GLYCEROL-PHOSPHONYL CHOLINE, 1-acyl-sn-glycerol-3-phosphate acyltransferase, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Robertson, R.M, Yao, J, Gajewski, S, Kumar, G, Martin, E.W, Rock, C.O, White, S.W. | | Deposit date: | 2016-07-21 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A two-helix motif positions the lysophosphatidic acid acyltransferase active site for catalysis within the membrane bilayer.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5LUH
 
 | | AadA E87Q in complex with ATP, calcium and streptomycin | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Stern, A.L, Van der Verren, S.E, Selmer, M. | | Deposit date: | 2016-09-08 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural mechanism of AadA, a dual-specificity aminoglycoside adenylyltransferase fromSalmonella enterica.
J.Biol.Chem., 293, 2018
|
|
6GKJ
 
 | | Translation initiation factor 4E in complex with trinucleotide mRNA 5' cap (m7GpppApG) | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4E, GLYCEROL | | Authors: | Warminski, M, Nowak, E, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2018-05-21 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.068 Å) | | Cite: | Translation initiation factor 4E in complex with trinucleotide mRNA 5' cap (m7GpppApG)
To Be Published
|
|
