7RCV
 
 | | High-resolution structure of photosystem II from the mesophilic cyanobacterium, Synechocystis sp. PCC 6803 | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Gisriel, C.J, Brudvig, G.W. | | Deposit date: | 2021-07-08 | | Release date: | 2021-12-29 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (2.01 Å) | | Cite: | High-resolution cryo-electron microscopy structure of photosystem II from the mesophilic cyanobacterium, Synechocystis sp. PCC 6803.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5CNC
 
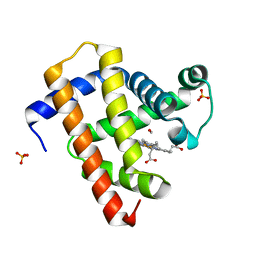 | | Ultrafast dynamics in myoglobin: 0.6 ps time delay | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Barends, T.R.M, Foucar, L, Ardevol, A, Nass, K.J, Aquila, A, Botha, S, Doak, R.B, Falahati, K, Hartmann, E, Hilpert, M, Heinz, M, Hoffmann, M.C, Koefinger, J, Koglin, J, Kovacsova, G, Liang, M, Milathianaki, D, Lemke, H.T, Reinstein, J, Roome, C.M, Shoeman, R.L, Williams, G.J, Burghardt, I, Hummer, G, Boutet, S, Schlichting, I. | | Deposit date: | 2015-07-17 | | Release date: | 2015-09-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Direct observation of ultrafast collective motions in CO myoglobin upon ligand dissociation.
Science, 350, 2015
|
|
5CRV
 
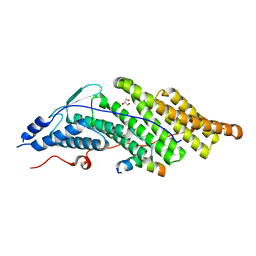 | | Crystal structure of the Bro domain of HD-PTP in a complex with the core region of STAM2 | | Descriptor: | GLYCEROL, Signal transducing adapter molecule 2, Tyrosine-protein phosphatase non-receptor type 23 | | Authors: | Lee, J, Ku, B, Kim, S.J. | | Deposit date: | 2015-07-23 | | Release date: | 2016-02-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural Study of the HD-PTP Bro1 Domain in a Complex with the Core Region of STAM2, a Subunit of ESCRT-0
Plos One, 11, 2016
|
|
7RM9
 
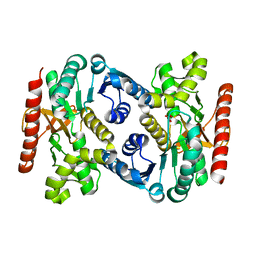 | | Human Malate Dehydrogenase I (MDHI) | | Descriptor: | MALONATE ION, Malate dehydrogenase, cytoplasmic | | Authors: | McCue, W, Finzel, B.C. | | Deposit date: | 2021-07-27 | | Release date: | 2022-01-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Characterization of the Human Cytosolic Malate Dehydrogenase I.
Acs Omega, 7, 2022
|
|
4X1S
 
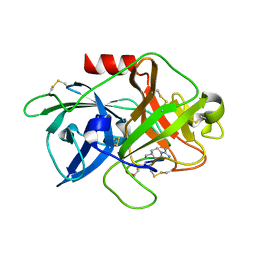 | | The crystal structure of mupain-1-16-D9A in complex with murinised human uPA at pH7.4 | | Descriptor: | Urokinase-type plasminogen activator, mupain-1-16, piperidine-1-carboximidamide | | Authors: | Jiang, L, Zhao, B, Xu, P, Andreasen, P, Huang, M. | | Deposit date: | 2014-11-25 | | Release date: | 2015-10-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A cyclic peptidic serine protease inhibitor: increasing affinity by increasing peptide flexibility.
Plos One, 9, 2014
|
|
8K1Y
 
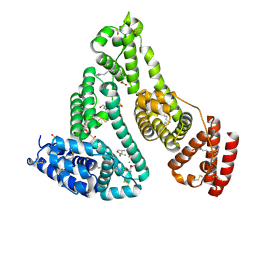 | | Crystal structure of human serum albumin and ruthenium Val complex adduct | | Descriptor: | Albumin, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Gong, W.J, Xie, L.L, Wang, W.M, Wang, H.F. | | Deposit date: | 2023-07-11 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of human serum albumin and ruthenium VaL complex adduct
To Be Published
|
|
7RLU
 
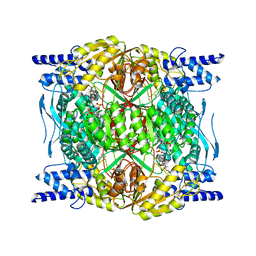 | | Structure of ALDH1L1 (10-formyltetrahydrofolate dehydrogenase) in complex with NADP | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Cytosolic 10-formyltetrahydrofolate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Tsybovsky, Y, Sereda, V, Golczak, M, Krupenko, N.I, Krupenko, S.A. | | Deposit date: | 2021-07-26 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of putative tumor suppressor ALDH1L1.
Commun Biol, 5, 2022
|
|
7RLT
 
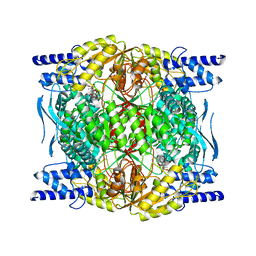 | | Structure of ligand-free ALDH1L1 (10-formyltetrahydrofolate dehydrogenase) | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Cytosolic 10-formyltetrahydrofolate dehydrogenase | | Authors: | Tsybovsky, Y, Sereda, V, Golczak, M, Krupenko, N.I, Krupenko, S.A. | | Deposit date: | 2021-07-26 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of putative tumor suppressor ALDH1L1.
Commun Biol, 5, 2022
|
|
4X0K
 
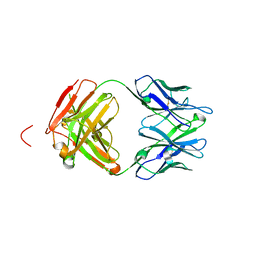 | | Engineered Fab fragment specific for EYMPME (EE) peptide | | Descriptor: | Fab fragment heavy chain, Fab fragment light chain | | Authors: | Johnson, J.L, Lieberman, R.L. | | Deposit date: | 2014-11-21 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural and biophysical characterization of an epitope-specific engineered Fab fragment and complexation with membrane proteins: implications for co-crystallization.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1YQL
 
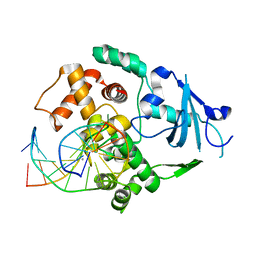 | | Catalytically inactive hOGG1 crosslinked with 7-deaza-8-azaguanine containing DNA | | Descriptor: | 5'-D(P*GP*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*C)-3', 5'-D(P*GP*TP*CP*CP*AP*(PPW)P*GP*TP*CP*TP*AP*C)-3', CALCIUM ION, ... | | Authors: | Banerjee, A, Yang, W, Karplus, M, Verdine, G.L. | | Deposit date: | 2005-02-02 | | Release date: | 2005-04-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA.
Nature, 434, 2005
|
|
7REH
 
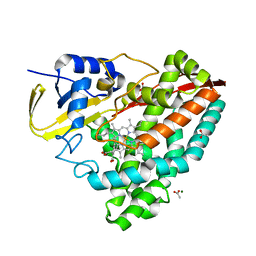 | |
7RKN
 
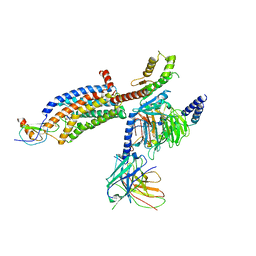 | | Structure of CX3CL1-US28-Gi-scFv16 in OC-state | | Descriptor: | Antibody fragment scFv16, Fractalkine, G-protein coupled receptor homolog US28, ... | | Authors: | Tsutsumi, N, Qu, Q, Jude, K.M, Skiniotis, G, Garcia, K.C. | | Deposit date: | 2021-07-22 | | Release date: | 2022-01-26 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Atypical structural snapshots of human cytomegalovirus GPCR interactions with host G proteins
Sci Adv, 8, 2022
|
|
7RKF
 
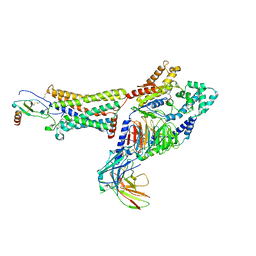 | | Structure of CX3CL1-US28-G11iN18-scFv16 in TL-state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody fragment scFv16, Fractalkine, ... | | Authors: | Tsutsumi, N, Maeda, S, Qu, Q, Skiniotis, G, Kobilka, B.K, Garcia, K.C. | | Deposit date: | 2021-07-22 | | Release date: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Atypical structural snapshots of human cytomegalovirus GPCR interactions with host G proteins
Sci Adv, 8, 2022
|
|
7RKM
 
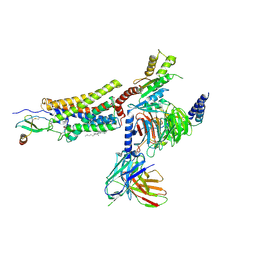 | | Structure of CX3CL1-US28-Gi-scFv16 in C-state | | Descriptor: | Antibody fragment scFv16, CHOLESTEROL, Fractalkine, ... | | Authors: | Tsutsumi, N, Qu, Q, Jude, K.M, Skiniotis, G, Garcia, K.C. | | Deposit date: | 2021-07-22 | | Release date: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Atypical structural snapshots of human cytomegalovirus GPCR interactions with host G proteins
Sci Adv, 8, 2022
|
|
7RRL
 
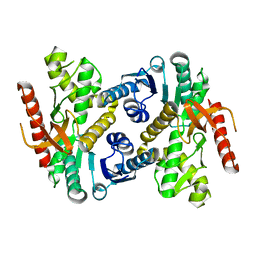 | |
7R76
 
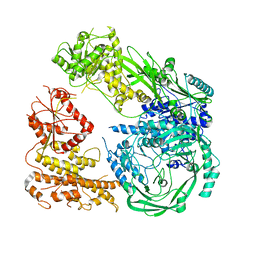 | | Cryo-EM structure of DNMT5 in apo state | | Descriptor: | DNA repair protein Rad8, ZINC ION | | Authors: | Wang, J, Patel, D.J. | | Deposit date: | 2021-06-24 | | Release date: | 2022-02-23 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into DNMT5-mediated ATP-dependent high-fidelity epigenome maintenance.
Mol.Cell, 82, 2022
|
|
5CRC
 
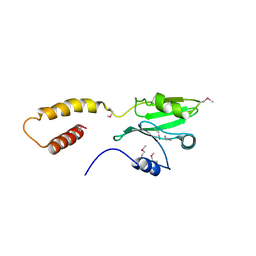 | | Structure of the SdeA DUB Domain | | Descriptor: | SdeA | | Authors: | Sheedlo, M.J, Qiu, J, Tan, Y, Paul, L.N, Luo, Z.Q, Das, C. | | Deposit date: | 2015-07-22 | | Release date: | 2015-11-25 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.853 Å) | | Cite: | Structural basis of substrate recognition by a bacterial deubiquitinase important for dynamics of phagosome ubiquitination.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7R77
 
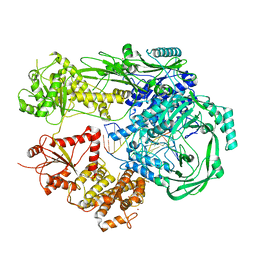 | |
7R78
 
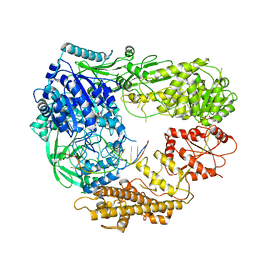 | |
7UM3
 
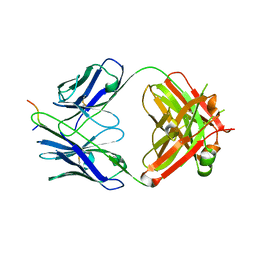 | | Crystal structure of a Fab in complex with a peptide derived from the LAG-3 D1 domain loop insertion | | Descriptor: | D1 domain loop peptide from Lymphocyte activation gene 3 protein, Fab heavy chain, Fab light chain | | Authors: | Zorn, J.A, Lee, P.S, Rajpal, A, Strop, P. | | Deposit date: | 2022-04-06 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3983 Å) | | Cite: | Preclinical Characterization of Relatlimab, a Human LAG-3-Blocking Antibody, Alone or in Combination with Nivolumab.
Cancer Immunol Res, 10, 2022
|
|
7RG8
 
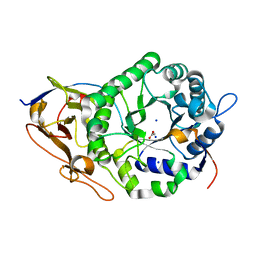 | | Crystal Structure of a Stable Heparanase Mutant | | Descriptor: | ACETATE ION, Heparanase 50 kDa subunit, Heparanase 8 kDa subunit, ... | | Authors: | Whitefield, C, Hong, N.S, Jackson, C.J. | | Deposit date: | 2021-07-14 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Computational design and experimental characterisation of a stable human heparanase variant.
Rsc Chem Biol, 3, 2022
|
|
4WY1
 
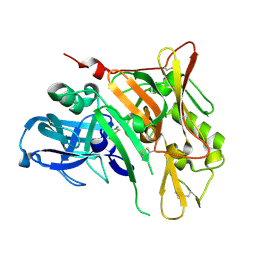 | | Crystal structure of human BACE-1 bound to Compound 24B | | Descriptor: | (4aR,8aS)-8a-(2,4-difluorophenyl)-4,4a,5,6,8,8a-hexahydropyrano[3,4-d][1,3]thiazin-2-amine, Beta-secretase 1 | | Authors: | Vajdos, F.F, Parris, K. | | Deposit date: | 2014-11-14 | | Release date: | 2015-03-04 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of a Series of Efficient, Centrally Efficacious BACE1 Inhibitors through Structure-Based Drug Design.
J.Med.Chem., 58, 2015
|
|
4LJ1
 
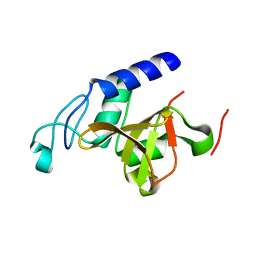 | | RipD (Rv1566c) from Mycobacterium tuberculosis: a non-catalytic NlpC/p60 domain protein with two penta-peptide repeat units (PVQQA-PVQPA) | | Descriptor: | Invasion-associated protein | | Authors: | Both, D, Steiner, E.M, Linder, D.C, Schnell, R, Schneider, G. | | Deposit date: | 2013-07-04 | | Release date: | 2013-11-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | RipD (Rv1566c) from Mycobacterium tuberculosis: adaptation of an NlpC/p60 domain to a non-catalytic peptidoglycan-binding function.
Biochem.J., 457, 2014
|
|
7R70
 
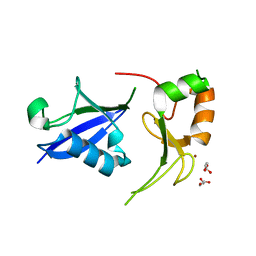 | | Crystal Structure of the UbArk2C fusion protein | | Descriptor: | GLYCEROL, Ubiquitin,E3 ubiquitin-protein ligase RNF165, ZINC ION | | Authors: | Paluda, A, Middleton, A.J, Mace, P.D, Day, C.L. | | Deposit date: | 2021-06-24 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Ubiquitin and a charged loop regulate the ubiquitin E3 ligase activity of Ark2C.
Nat Commun, 13, 2022
|
|
7R71
 
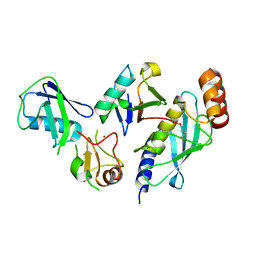 | | Crystal Structure of the UbArk2C-UbcH5b~Ub complex | | Descriptor: | Ubiquitin, Ubiquitin,E3 ubiquitin-protein ligase RNF165, Ubiquitin-conjugating enzyme E2 D2, ... | | Authors: | Paluda, A, Middleton, A.J, Mace, P.D, Day, C.L. | | Deposit date: | 2021-06-24 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ubiquitin and a charged loop regulate the ubiquitin E3 ligase activity of Ark2C.
Nat Commun, 13, 2022
|
|
