3ILJ
 
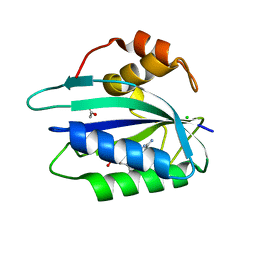 | | Crystal structure of E. coli HPPK(D95A) in complex with MgAMPCPP | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2009-08-07 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and functional roles of residues D95 and D97 in E. coli HPPK
To be Published
|
|
5K54
 
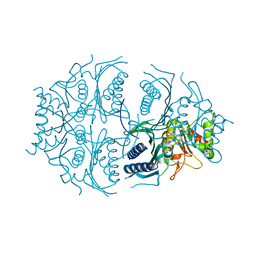 | | Human muscle fructose-1,6-bisphosphatase E69Q mutant in active R-state | | Descriptor: | Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2016-05-23 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | Structural studies of human muscle FBPase
To Be Published
|
|
3MHR
 
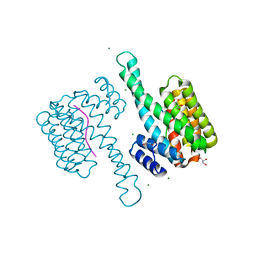 | | 14-3-3 sigma in complex with YAP pS127-peptide | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Schumacher, B, Skwarczynska, M, Rose, R, Ottmann, C. | | Deposit date: | 2010-04-08 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure of a 14-3-3[sigma]-YAP phosphopeptide complex at 1.15 A resolution
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2WDW
 
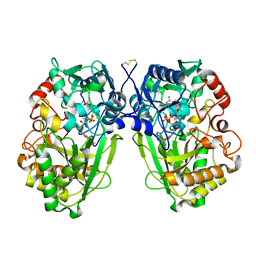 | | The Native Crystal Structure of the Primary Hexose Oxidase (Dbv29) in Antibiotic A40926 Biosynthesis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PUTATIVE HEXOSE OXIDASE | | Authors: | Liu, Y.-C, Li, Y.-S, Lyu, S.-Y, Chen, Y.-H, Chan, H.-C, Huang, C.-J, Huang, Y.-T, Chen, G.-H, Chou, C.-C, Tsai, M.-D, Li, T.-L. | | Deposit date: | 2009-03-27 | | Release date: | 2010-04-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Interception of Teicoplanin Oxidation Intermediates Yields New Antimicrobial Scaffolds
Nat.Chem.Biol., 7, 2011
|
|
3SWK
 
 | |
3GD3
 
 | |
6KO6
 
 | | Crystal structure of AMPPNP bound Cka1 from C. neoformans | | Descriptor: | CMGC/CK2 protein kinase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Cho, H.S, Yoo, Y. | | Deposit date: | 2019-08-08 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analysis of fungal pathogenicity-related casein kinase alpha subunit, Cka1, in the human fungal pathogen Cryptococcus neoformans.
Sci Rep, 9, 2019
|
|
5TZ1
 
 | | Crystal structure of sterol 14-alpha demethylase (CYP51) from Candida albicans in complex with the tetrazole-based antifungal drug candidate VT1161 (VT1) | | Descriptor: | (R)-2-(2,4-Difluorophenyl)-1,1-difluoro-3-(1H-tetrazol-1-yl)-1-(5-(4-(2,2,2-trifluoroethoxy)phenyl)pyridin-2-yl)propan-2-ol, PROTOPORPHYRIN IX CONTAINING FE, Sterol 14-alpha demethylase | | Authors: | Hargrove, T, Wawrzak, Z, Lepesheva, G. | | Deposit date: | 2016-11-21 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analyses of Candida albicans sterol 14 alpha-demethylase complexed with azole drugs address the molecular basis of azole-mediated inhibition of fungal sterol biosynthesis.
J. Biol. Chem., 292, 2017
|
|
2WTY
 
 | | Crystal structure of the homodimeric MafB in complex with the T-MARE binding site | | Descriptor: | DNA (5'-D(*TP*AP*AP*TP*TP*GP*CP*TP*GP*AP*CP*TP*CP*AP *GP*CP*AP*AP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*TP*TP*GP*CP*TP*GP*AP*GP*TP*CP*AP *GP*CP*AP*AP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Consani Textor, L, Holton, S, Wilmanns, M. | | Deposit date: | 2009-09-25 | | Release date: | 2010-12-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design of a bZIP Transcription Factor with Homo/Heterodimer-Induced DNA-Binding Preference.
Structure, 22, 2014
|
|
3DRM
 
 | | 2.2 Angstrom Crystal Structure of Thr114Phe Alpha1-Antitrypsin | | Descriptor: | Alpha-1-antitrypsin | | Authors: | Gooptu, B, Nobeli, I, Purkiss, A, Phillips, R.L, Mallya, M, Lomas, D.A, Barrett, T.E. | | Deposit date: | 2008-07-11 | | Release date: | 2009-03-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic and cellular characterisation of two mechanisms stabilising the native fold of alpha1-antitrypsin: implications for disease and drug design.
J.Mol.Biol., 387, 2009
|
|
3BGC
 
 | | HIV-1 protease in complex with a benzyl decorated oligoamine | | Descriptor: | CHLORIDE ION, N,N'-(iminodiethane-2,1-diyl)bis(4-amino-N-benzylbenzenesulfonamide), Protease | | Authors: | Boettcher, J, Blum, A, Sammet, B, Heine, A, Diederich, W.E, Klebe, G. | | Deposit date: | 2007-11-26 | | Release date: | 2008-09-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Achiral oligoamines as versatile tool for the development of aspartic protease inhibitors
Bioorg.Med.Chem., 16, 2008
|
|
1CLF
 
 | | CLOSTRIDIUM PASTEURIANUM FERREDOXIN | | Descriptor: | FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Bertini, I, Donaire, A, Feinberg, B.A, Luchinat, C, Piccioli, M, Yuan, H. | | Deposit date: | 1995-06-21 | | Release date: | 1996-01-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the oxidized 2[4Fe-4S] ferredoxin from Clostridium pasteurianum.
Eur.J.Biochem., 232, 1995
|
|
5UW5
 
 | | PCY1 H695A Variant in Complex with Follower Peptide | | Descriptor: | CACODYLATE ION, CALCIUM ION, Peptide cyclase 1, ... | | Authors: | Chekan, J.R, Nair, S.K. | | Deposit date: | 2017-02-20 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Characterization of the macrocyclase involved in the biosynthesis of RiPP cyclic peptides in plants.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3S7K
 
 | | Structure of thrombin mutant Y225P in the E form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, POTASSIUM ION, Prothrombin | | Authors: | Niu, W, Chen, Z, Gandhi, P, Vogt, A, Pozzi, N, Pele, L.A, Zapata, F, Di Cera, E. | | Deposit date: | 2011-05-26 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic and Kinetic Evidence of Allostery in a Trypsin-like Protease.
Biochemistry, 50, 2011
|
|
5CEL
 
 | | CBH1 (E212Q) CELLOTETRAOSE COMPLEX | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, COBALT (II) ION, ... | | Authors: | Divne, C, Stahlberg, J, Jones, T.A. | | Deposit date: | 1997-09-24 | | Release date: | 1997-12-24 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution crystal structures reveal how a cellulose chain is bound in the 50 A long tunnel of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 275, 1998
|
|
2Y92
 
 | | Crystal structure of MAL adaptor protein | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, TOLL/INTERLEUKIN-1 RECEPTOR DOMAIN-CONTAINING ADAPTER PROTEIN, | | Authors: | Valkov, E, Stamp, A, Martin, J.L, Kobe, B. | | Deposit date: | 2011-02-11 | | Release date: | 2011-09-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal Structure of Toll-Like Receptor Adaptor Mal/Tirap Reveals the Molecular Basis for Signal Transduction and Disease Protection.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6ANQ
 
 | | STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING D4TTP AT PH 8.5 | | Descriptor: | 2',3'-DEHYDRO-2',3'-DEOXY-THYMIDINE 5'-TRIPHOSPHATE, DNA PRIMER (5'- D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*GP)-3'), DNA TEMPLATE (5'- D(*AP*TP*GP*AP*AP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), ... | | Authors: | Martinez, S.E, Das, K, Arnold, E. | | Deposit date: | 2017-08-14 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.586 Å) | | Cite: | Structure of HIV-1 reverse transcriptase/d4TTP complex: Novel DNA cross-linking site and pH-dependent conformational changes.
Protein Sci., 28, 2019
|
|
2P8W
 
 | | Fitted structure of eEF2 in the 80S:eEF2:GDPNP cryo-EM reconstruction | | Descriptor: | Elongation factor 2, Elongation factor Tu-B, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Taylor, D.J, Nilsson, J, Merrill, A.R, Andersen, G.R, Nissen, P, Frank, J. | | Deposit date: | 2007-03-23 | | Release date: | 2007-05-08 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (11.3 Å) | | Cite: | Structures of modified eEF2.80S ribosome complexes reveal the role of GTP hydrolysis in translocation.
Embo J., 26, 2007
|
|
3KD5
 
 | |
2YQ0
 
 | |
2YPZ
 
 | |
1NOY
 
 | | DNA POLYMERASE (E.C.2.7.7.7)/DNA COMPLEX | | Descriptor: | DNA (5'-D(*TP*TP*T)-3'), MANGANESE (II) ION, PROTEIN (DNA POLYMERASE (E.C.2.7.7.7)), ... | | Authors: | Wang, J, Yu, P, Lin, T.C, Konigsberg, W.H, Steitz, T.A. | | Deposit date: | 1996-02-16 | | Release date: | 1996-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of an NH2-terminal fragment of T4 DNA polymerase and its complexes with single-stranded DNA and with divalent metal ions.
Biochemistry, 35, 1996
|
|
3GGV
 
 | | HIV Protease, pseudo-symmetric inhibitors | | Descriptor: | V-1 protease, methyl [(1S)-1-{[(1R,3S,4S)-3-hydroxy-4-{[(2S)-2-(3-{[6-(1-hydroxy-1-methylethyl)pyridin-2-yl]methyl}-2-oxo-2,3-dihydro-1H-imidazol-1-yl)-3,3-dimethylbutanoyl]amino}-5-phenyl-1-(4-pyridin-2-ylbenzyl)pentyl]carbamoyl}-2,2-dimethylpropyl]carbamate | | Authors: | Stoll, V.S. | | Deposit date: | 2009-03-02 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | 2-Pyridyl P1'-substituted symmetry-based human immunodeficiency virus
protease inhibitors (A-792611 and A-790742) with potential for convenient
dosing and reduced side effects.
J.Med.Chem., 52, 2009
|
|
2YPY
 
 | |
2WOF
 
 | | EDTA treated E. coli copper amine oxidase | | Descriptor: | COPPER (II) ION, PRIMARY AMINE OXIDASE, SODIUM ION | | Authors: | Smith, M.A, Pirrat, P, Pearson, A.R, Knowles, P.F, Phillips, S.E.V, McPherson, M.J. | | Deposit date: | 2009-07-23 | | Release date: | 2010-05-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Exploring the Roles of the Metal Ions in Escherichia Coli Copper Amine Oxidase.
Biochemistry, 49, 2010
|
|
