1OUH
 
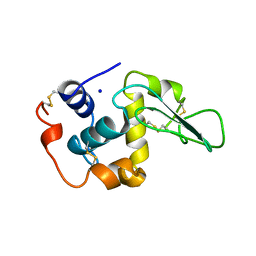 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: X-RAY STRUCTURE OF THE V74A MUTANT | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Fujii, S, Yutani, K. | | Deposit date: | 1996-08-23 | | Release date: | 1997-02-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of the hydrophobic effect to the stability of human lysozyme: calorimetric studies and X-ray structural analyses of the nine valine to alanine mutants.
Biochemistry, 36, 1997
|
|
3PJ8
 
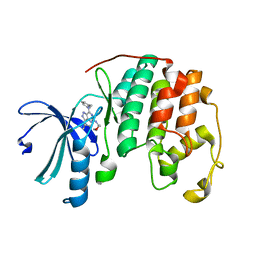 | | Structure of CDK2 in complex with a Pyrazolo[4,3-d]pyrimidine Bioisostere of Roscovitine. | | Descriptor: | (2R)-2-{[7-(benzylamino)-3-(propan-2-yl)-1H-pyrazolo[4,3-d]pyrimidin-5-yl]amino}butan-1-ol, Cell division protein kinase 2 | | Authors: | McNae, I.W, Jorda, R, Havlicek, L, Strnad, M, Voller, J, Walkinshaw, M.D, Krystof, V. | | Deposit date: | 2010-11-09 | | Release date: | 2011-04-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Pyrazolo[4,3-d]pyrimidine Bioisostere of Roscovitine: Evaluation of a Novel Selective Inhibitor of Cyclin-Dependent Kinases with Antiproliferative Activity.
J.Med.Chem., 54, 2011
|
|
4RXY
 
 | |
1OUF
 
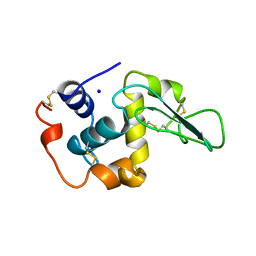 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: X-RAY STRUCTURE OF THE V130A MUTANT | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Fujii, S, Yutani, K. | | Deposit date: | 1996-08-23 | | Release date: | 1997-02-12 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of the hydrophobic effect to the stability of human lysozyme: calorimetric studies and X-ray structural analyses of the nine valine to alanine mutants.
Biochemistry, 36, 1997
|
|
4RKK
 
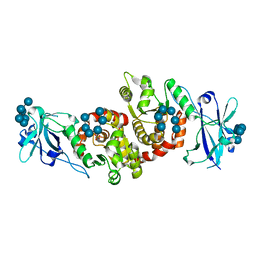 | | Structure of a product bound phosphatase | | Descriptor: | Laforin, PHOSPHATE ION, alpha-D-glucopyranose, ... | | Authors: | Vander Kooi, C.W. | | Deposit date: | 2014-10-13 | | Release date: | 2015-01-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural mechanism of laforin function in glycogen dephosphorylation and lafora disease.
Mol.Cell, 57, 2015
|
|
4RPX
 
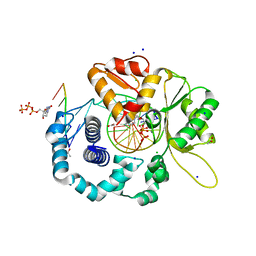 | |
1O0C
 
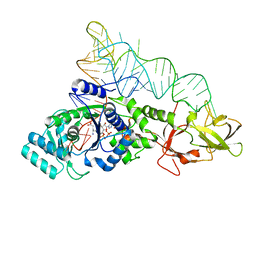 | |
1AHT
 
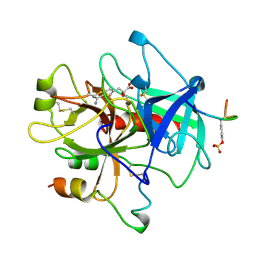 | |
5KP7
 
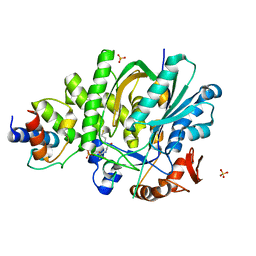 | |
1O3Z
 
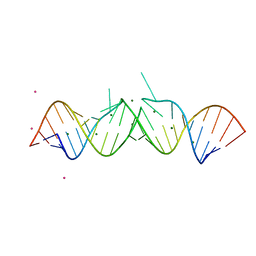 | | HIV-1 DIS(MAL) DUPLEX RU HEXAMINE-SOAKED | | Descriptor: | HIV-1 DIS(MAL) GENOMIC RNA, MAGNESIUM ION, RUTHENIUM ION | | Authors: | Ennifar, E, Walter, P, Dumas, P. | | Deposit date: | 2003-05-16 | | Release date: | 2003-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A crystallographic study of the binding of 13 metal ions to two related RNA duplexes.
Nucleic Acids Res., 31, 2003
|
|
1HDT
 
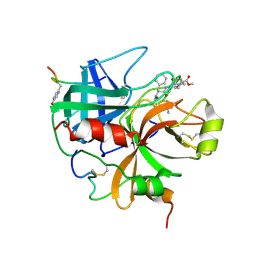 | |
2PUQ
 
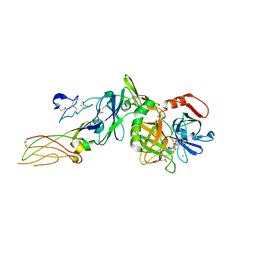 | |
1BI9
 
 | | RETINAL DEHYDROGENASE TYPE TWO WITH NAD BOUND | | Descriptor: | CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, RETINAL DEHYDROGENASE TYPE II | | Authors: | Newcomer, M.E, Lamb, A.L. | | Deposit date: | 1998-06-23 | | Release date: | 1999-07-22 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of retinal dehydrogenase type II at 2.7 A resolution: implications for retinal specificity.
Biochemistry, 38, 1999
|
|
4TGF
 
 | | SOLUTION STRUCTURES OF HUMAN TRANSFORMING GROWTH FACTOR ALPHA DERIVED FROM 1*H NMR DATA | | Descriptor: | DES-VAL-1,VAL-2,TRANSFORMING GROWTH FACTOR, ALPHA | | Authors: | Kline, T.P, Brown, F.K, Brown, S.C, Jeffs, P.W, Kopple, K.D, Mueller, L. | | Deposit date: | 1990-06-13 | | Release date: | 1991-10-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of human transforming growth factor alpha derived from 1H NMR data.
Biochemistry, 29, 1990
|
|
8R8D
 
 | |
6UOM
 
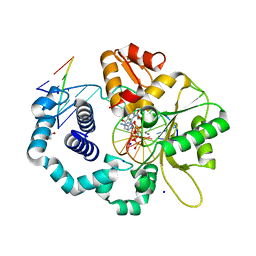 | | Y271G DNA polymerase beta ternary complex with templating adenine and incoming r8-oxo-GTP | | Descriptor: | 1,2-ETHANEDIOL, 2',3'-DIDEOXYCYTIDINE-5'-MONOPHOSPHATE, 8-OXO-GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Smith, M.R, Freudenthal, B.D. | | Deposit date: | 2019-10-15 | | Release date: | 2020-01-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Molecular and structural characterization of oxidized ribonucleotide insertion into DNA by human DNA polymerase beta.
J.Biol.Chem., 295, 2020
|
|
1FIW
 
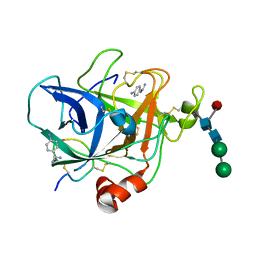 | | THREE-DIMENSIONAL STRUCTURE OF BETA-ACROSIN FROM RAM SPERMATOZOA | | Descriptor: | BETA-ACROSIN HEAVY CHAIN, BETA-ACROSIN LIGHT CHAIN, P-AMINO BENZAMIDINE, ... | | Authors: | Tranter, R, Read, J.A, Jones, R, Brady, R.L. | | Deposit date: | 2000-08-07 | | Release date: | 2000-11-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Effector sites in the three-dimensional structure of mammalian sperm beta-acrosin.
Structure Fold.Des., 8, 2000
|
|
7MKF
 
 | |
1R4M
 
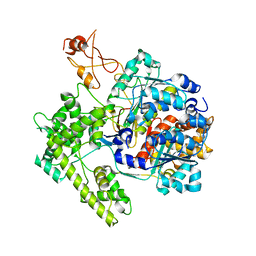 | | APPBP1-UBA3-NEDD8, an E1-ubiquitin-like protein complex | | Descriptor: | Ubiquitin-like protein NEDD8, ZINC ION, amyloid beta precursor protein-binding protein 1, ... | | Authors: | Walden, H, Podgorski, M.S, Holton, J.M, Schulman, B.A. | | Deposit date: | 2003-10-07 | | Release date: | 2003-12-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the APPBP1-UBA3-NEDD8-ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1.
Mol.Cell, 12, 2003
|
|
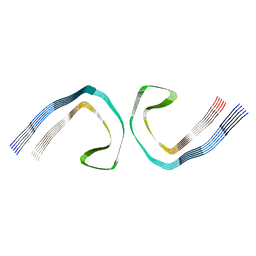
7MKH
 
 | |
7MKG
 
 | |
5JUM
 
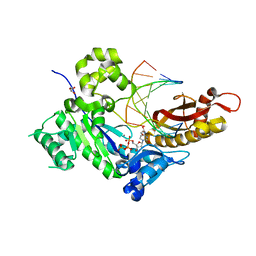 | | Crystal Structure of Human DNA Polymerase Eta Inserting dCTP Opposite N-(2'-deoxyguanosin-8- yl)-3-aminobenzanthrone (C8-dG-ABA) | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), ... | | Authors: | Patra, A, Politica, D.A, Stone, M.P, Egli, M. | | Deposit date: | 2016-05-10 | | Release date: | 2016-09-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism of Error-Free Bypass of the Environmental Carcinogen N-(2'-Deoxyguanosin-8-yl)-3-aminobenzanthrone Adduct by Human DNA Polymerase eta.
Chembiochem, 17, 2016
|
|
5L3K
 
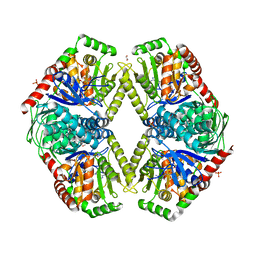 | | Structure of Mycobacterium thermoresistibile trehalose-6-phosphate synthase in a ternary complex with ADP and fructose-6-phosphate | | Descriptor: | 1,2-ETHANEDIOL, 6-O-phosphono-beta-D-fructofuranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Mendes, V, Verma, N, Blaszczyk, M, Blundell, T.L. | | Deposit date: | 2016-05-23 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.305 Å) | | Cite: | Mycobacterial OtsA Structures Unveil Substrate Preference Mechanism and Allosteric Regulation by 2-Oxoglutarate and 2-Phosphoglycerate.
Mbio, 10, 2019
|
|
1PLF
 
 | |
7N0H
 
 | | CryoEM structure of SARS-CoV-2 spike protein (S-6P, 2-up) in complex with sybodies (Sb45) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Jiang, J, Huang, R, Margulies, D. | | Deposit date: | 2021-05-25 | | Release date: | 2021-06-02 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structures of synthetic nanobody-SARS-CoV-2 receptor-binding domain complexes reveal distinct sites of interaction.
J.Biol.Chem., 297, 2021
|
|
