2ONE
 
 | |
1GVD
 
 | | CRYSTAL STRUCTURE OF C-MYB R2 V103L MUTANT | | Descriptor: | AMMONIUM ION, MYB PROTO-ONCOGENE PROTEIN, SULFATE ION | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2002-02-08 | | Release date: | 2003-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of C-Myb DNA-Binding Domain: Specific Na+ Binding and Correlation with NMR Structure
To be Published
|
|
1H9P
 
 | | Crystal Structure of Dioclea guianensis Seed Lectin | | Descriptor: | CADMIUM ION, LECTIN ALPHA CHAIN, MANGANESE (II) ION | | Authors: | Romero, A, Wah, D.A, Gallego Del sol, F, Cavada, B.S, Ramos, M.V, Grangeiro, T.B, Sampaio, A.H, Calvete, J.J. | | Deposit date: | 2001-03-16 | | Release date: | 2001-03-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Native and Cd/Cd-Substituted Dioclea Guianensis Seed Lectin. A Novel Manganese-Binding Site and Structural Basis of Dimer-Tetramer Association
J.Mol.Biol., 310, 2001
|
|
6H2L
 
 | |
5NJ6
 
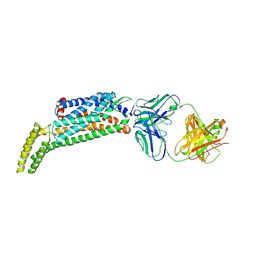 | | Crystal structure of a thermostabilised human protease-activated receptor-2 (PAR2) in ternary complex with Fab3949 and AZ7188 at 4.0 angstrom resolution | | Descriptor: | Fab3949 H, Fab3949 L, Proteinase-activated receptor 2,Soluble cytochrome b562,Proteinase-activated receptor 2 | | Authors: | Cheng, R.K.Y, Fiez-Vandal, C, Schlenker, O, Edman, K, Aggeler, B, Brown, D.G, Brown, G, Cooke, R.M, Dumelin, C.E, Dore, A.S, Geschwindner, S, Grebner, C, Hermansson, N.-O, Jazayeri, A, Johansson, P, Leong, L, Prihandoko, R, Rappas, M, Soutter, H, Snijder, A, Sundstrom, L, Tehan, B, Thornton, P, Troast, D, Wiggin, G, Zhukov, A, Marshall, F.H, Dekker, N. | | Deposit date: | 2017-03-28 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural insight into allosteric modulation of protease-activated receptor 2.
Nature, 545, 2017
|
|
3KEV
 
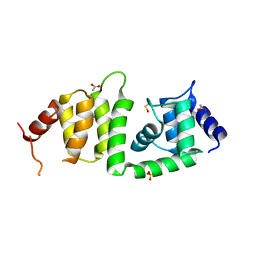 | | X-ray crystal structure of a DCUN1 domain-containing protein from Galdieria sulfuraria | | Descriptor: | ACETATE ION, Galieria sulfuraria DCUN1 domain-containing protein, SULFATE ION | | Authors: | Burgie, E.S, Bingman, C.A, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-10-26 | | Release date: | 2009-12-01 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural architecture of Galdieria sulphuraria DCN1L.
Proteins, 79, 2011
|
|
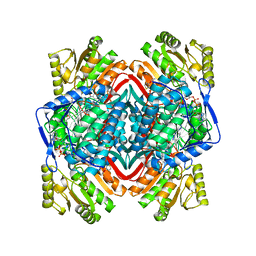
6B5G
 
 | |
1LYB
 
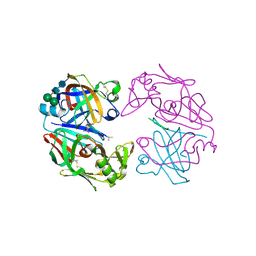 | | CRYSTAL STRUCTURES OF NATIVE AND INHIBITED FORMS OF HUMAN CATHEPSIN D: IMPLICATIONS FOR LYSOSOMAL TARGETING AND DRUG DESIGN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CATHEPSIN D, PEPSTATIN, ... | | Authors: | Baldwin, E.T, Bhat, T.N, Gulnik, S, Erickson, J.W. | | Deposit date: | 1993-04-22 | | Release date: | 1994-01-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of native and inhibited forms of human cathepsin D: implications for lysosomal targeting and drug design.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
5LSK
 
 | | CRYSTAL STRUCTURE OF THE HUMAN KINETOCHORE MIS12-CENP-C COMPLEX | | Descriptor: | Centromere protein C, Kinetochore-associated protein DSN1 homolog, Kinetochore-associated protein NSL1 homolog, ... | | Authors: | Vetter, I.R, Petrovic, A, Keller, J, Liu, Y. | | Deposit date: | 2016-09-02 | | Release date: | 2016-11-16 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (3.502 Å) | | Cite: | Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores.
Cell, 167, 2016
|
|
7YC2
 
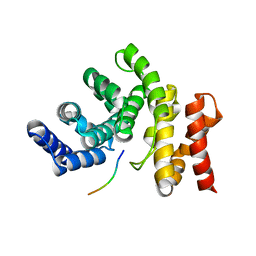 | |
1HCC
 
 | |
5MKJ
 
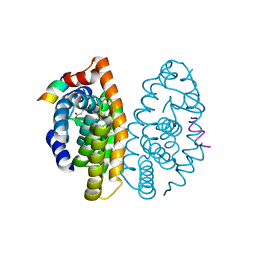 | | Crystal structure of the Retinoid X Receptor alpha in complex with synthetic honokiol derivative 9 and a fragment of the TIF2 co-activator. | | Descriptor: | (~{E})-3-[4-oxidanyl-3-(5-prop-2-enyl-2-propoxy-phenyl)phenyl]prop-2-enoic acid, LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN-ASP, Retinoic acid receptor RXR-alpha | | Authors: | Andrei, S.A, Brunsveld, L, Scheepstra, M, Ottmann, C. | | Deposit date: | 2016-12-05 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand Dependent Switch from RXR Homo- to RXR-NURR1 Heterodimerization.
ACS Chem Neurosci, 8, 2017
|
|
2END
 
 | | CRYSTAL STRUCTURE OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME FROM BACTERIOPHAGE T4: REFINEMENT AT 1.45 ANGSTROMS AND X-RAY ANALYSIS OF THE THREE ACTIVE SITE MUTANTS | | Descriptor: | ENDONUCLEASE V | | Authors: | Vassylyev, D.G, Ariyoshi, M, Matsumoto, O, Katayanagi, K, Ohtsuka, E, Morikawa, K. | | Deposit date: | 1994-08-08 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a pyrimidine dimer-specific excision repair enzyme from bacteriophage T4: refinement at 1.45 A and X-ray analysis of the three active site mutants.
J.Mol.Biol., 249, 1995
|
|
6B5I
 
 | |
8H5B
 
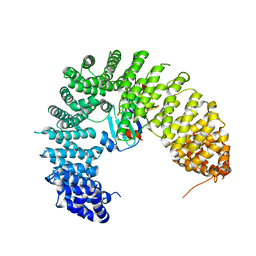 | | The cryo-EM structure of nuclear transport receptor Kap114p complex with yeast TATA-box binding protein | | Descriptor: | Importin subunit beta-5, TATA-box-binding protein | | Authors: | Hsia, K.C, Liao, C.C, Wang, C.H, Wu, Y.M. | | Deposit date: | 2022-10-12 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Structural convergence endows nuclear transport receptor Kap114p with a transcriptional repressor function toward TATA-binding protein.
Nat Commun, 14, 2023
|
|
5NN6
 
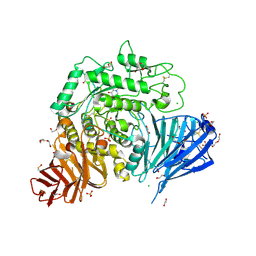 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with N-hydroxyethyl-1-deoxynojirimycin | | Descriptor: | (2R,3R,4R,5S)-1-(2-hydroxyethyl)-2-(hydroxymethyl)piperidine-3,4,5-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roig-Zamboni, V, Cobucci-Ponzano, B, Iacono, R, Ferrara, M.C, Germany, S, Parenti, G, Bourne, Y, Moracci, M. | | Deposit date: | 2017-04-08 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human lysosomal acid alpha-glucosidase-a guide for the treatment of Pompe disease.
Nat Commun, 8, 2017
|
|
5NN4
 
 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with N-acetyl-cysteine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roig-Zamboni, V, Cobucci-Ponzano, B, Iacono, R, Ferrara, M.C, Germany, S, Parenti, G, Bourne, Y, Moracci, M. | | Deposit date: | 2017-04-08 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of human lysosomal acid alpha-glucosidase-a guide for the treatment of Pompe disease.
Nat Commun, 8, 2017
|
|
7WJU
 
 | | Cryo-EM structure of the AsCas12f1-sgRNAv1-dsDNA ternary complex | | Descriptor: | Non-target strand, OrfB_Zn_ribbon domain-containing protein, Target strand, ... | | Authors: | Wu, Z, Liu, D, Shen, H, Ji, Q. | | Deposit date: | 2022-01-07 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structure-directed functional evolution of the miniature CRISPR-AsCas12f1 system
To Be Published
|
|
1HE1
 
 | | Crystal structure of the complex between the GAP domain of the Pseudomonas aeruginosa ExoS toxin and human Rac | | Descriptor: | ALUMINUM FLUORIDE, EXOENZYME S, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wurtele, M, Wolf, E, Pederson, K.J, Buchwald, G, Ahmadian, M.R, Barbieri, J.T, Wittinghofer, A. | | Deposit date: | 2000-11-18 | | Release date: | 2001-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How the Pseudomonas Aeruginosa Exos Toxin Downregulates Rac
Nat.Struct.Biol., 8, 2001
|
|
5NKX
 
 | | HRSV M2-1 core domain, P3221 crystal form | | Descriptor: | M2-1 | | Authors: | Almeida Hernandez, Y, Josts, I, Molina, I.G, de Pray-Gay, G, Tidow, H. | | Deposit date: | 2017-04-03 | | Release date: | 2018-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.00008678 Å) | | Cite: | Structure and stability of the Human respiratory syncytial virus M2-1RNA-binding core domain reveals a compact and cooperative folding unit.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
2CI2
 
 | |
3KDF
 
 | | X-ray Crystal Structure of the Human Replication Protein A Complex from Wheat Germ Cell Free Expression | | Descriptor: | 1,2-ETHANEDIOL, Replication protein A 14 kDa subunit, Replication protein A 32 kDa subunit | | Authors: | Burgie, E.S, Bingman, C.A, Phillips Jr, G.N, Fox, B.G, Makino, S.-I, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-10-22 | | Release date: | 2009-12-01 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (1.975 Å) | | Cite: | X-ray Crystal Structure of the Human Replication Protein A Complex from Wheat Germ Cell Free Expression
To be Published
|
|
6BML
 
 | | Structure of human DHHC20 palmitoyltransferase, irreversibly inhibited by 2-bromopalmitate | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-DIPHOSPHATE, PALMITIC ACID, PHOSPHATE ION, ... | | Authors: | Rana, M.S, Lee, C.-J, Banerjee, A. | | Deposit date: | 2017-11-15 | | Release date: | 2018-01-24 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Fatty acyl recognition and transfer by an integral membraneS-acyltransferase.
Science, 359, 2018
|
|
3NIB
 
 | | Teg14 Apo | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Teg14 | | Authors: | Bick, M.J, Banik, J.J, Darst, S.A, Brady, S.F. | | Deposit date: | 2010-06-15 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The 2.7 A resolution structure of the glycopeptide sulfotransferase Teg14
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2CBS
 
 | | CELLULAR RETINOIC ACID BINDING PROTEIN II IN COMPLEX WITH A SYNTHETIC RETINOIC ACID (RO-13 6307) | | Descriptor: | 3-METHYL-7-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRO-NAPHTHALEN-2-YL) -OCTA-2,4,6-TRIENOIC ACID, PROTEIN (CRABP-II) | | Authors: | Chaudhuri, B, Kleywegt, G.J, Bergfors, T, Jones, T.A. | | Deposit date: | 1999-02-22 | | Release date: | 1999-12-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of cellular retinoic acid binding proteins I and II in complex with synthetic retinoids.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
