3GYT
 
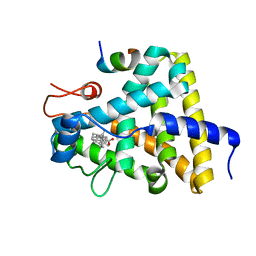 | | Nuclear receptor DAF-12 from parasitic nematode Strongyloides stercoralis in complex with its physiological ligand dafachronic acid delta 4 | | Descriptor: | (14beta,17alpha,25R)-3-oxocholest-4-en-26-oic acid, Nuclear hormone receptor of the steroid/thyroid hormone receptors superfamily, SRC1 | | Authors: | Zhou, X.E, Wang, Z, Suino-Powell, K, Motola, D.L, Conneely, A, Ogata, C, Sharma, K.K, Auchus, R.J, Kliewer, S.A, Xu, H.E, Mangelsdorf, D.J. | | Deposit date: | 2009-04-05 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of the nuclear receptor DAF-12 as a therapeutic target in parasitic nematodes.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3H8G
 
 | | Bestatin complex structure of leucine aminopeptidase from Pseudomonas putida | | Descriptor: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, BICARBONATE ION, Cytosol aminopeptidase, ... | | Authors: | Kale, A, Dijkstra, B.W, Sonke, T, Thunnissen, A.M.W.H. | | Deposit date: | 2009-04-29 | | Release date: | 2010-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the leucine aminopeptidase from Pseudomonas putida reveals the molecular basis for its enantioselectivity and broad substrate specificity.
J.Mol.Biol., 398, 2010
|
|
3H0G
 
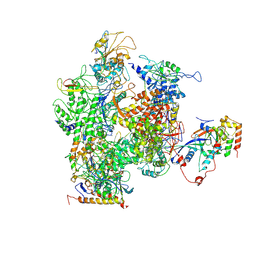 | | RNA Polymerase II from Schizosaccharomyces pombe | | Descriptor: | DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Spahr, H, Calero, G, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2009-04-09 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Schizosacharomyces pombe RNA polymerase II at 3.6-A resolution.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3H3Z
 
 | |
3HAF
 
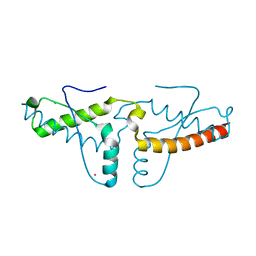 | | Human prion protein variant V129 domain swapped dimer | | Descriptor: | CADMIUM ION, CHLORIDE ION, Major prion protein | | Authors: | Lee, S, Antony, L, Hartmann, R, Knaus, K.J, Surewicz, K, Surewicz, W.K, Yee, V.C. | | Deposit date: | 2009-05-01 | | Release date: | 2010-01-12 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Conformational diversity in prion protein variants influences intermolecular beta-sheet formation.
Embo J., 29, 2010
|
|
3H4J
 
 | | crystal structure of pombe AMPK KDAID fragment | | Descriptor: | SNF1-like protein kinase ssp2 | | Authors: | Chen, L, Jiao, Z.-H, Zheng, L.-S, Zhang, Y.-Y, Xie, S.-T, Wang, Z.-X, Wu, J.-W. | | Deposit date: | 2009-04-20 | | Release date: | 2009-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural insight into the autoinhibition mechanism of AMP-activated protein kinase
Nature, 459, 2009
|
|
3H4X
 
 | |
3H6V
 
 | | Crystal structure of the iGluR2 ligand-binding core (S1S2J-N754S) in complex with glutamate and NS5206 at 2.10 A resolution | | Descriptor: | (3R)-3-cyclopentyl-7-[(4-methylpiperazin-1-yl)sulfonyl]-3,4-dihydro-2H-1,2-benzothiazine 1,1-dioxide, DIMETHYL SULFOXIDE, GLUTAMIC ACID, ... | | Authors: | Hald, H, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2009-04-24 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Distinct structural features of cyclothiazide are responsible for effects on peak current amplitude and desensitization kinetics at iGluR2.
J.Mol.Biol., 391, 2009
|
|
3H6Z
 
 | |
3H7B
 
 | |
3H7O
 
 | |
3H7T
 
 | |
3H8W
 
 | |
3HAK
 
 | | Human prion protein variant V129 | | Descriptor: | Major prion protein | | Authors: | Lee, S, Antony, L, Hartmann, R, Knaus, K.J, Surewicz, K, Surewicz, W.K, Yee, V.C. | | Deposit date: | 2009-05-01 | | Release date: | 2010-01-12 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformational diversity in prion protein variants influences intermolecular beta-sheet formation.
Embo J., 29, 2010
|
|
3H8U
 
 | |
3HB2
 
 | | PrtC methionine mutants: M226I | | Descriptor: | CALCIUM ION, CHLORIDE ION, Secreted protease C, ... | | Authors: | Oberholzer, A.E, Bumann, M, Hege, T, Russo, S, Baumann, U. | | Deposit date: | 2009-05-04 | | Release date: | 2009-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Metzincin's canonical methionine is responsible for the structural integrity of the zinc-binding site
Biol.Chem., 390, 2009
|
|
3H8Z
 
 | | The Crystal Structure of the Tudor Domains from FXR2 | | Descriptor: | Fragile X mental retardation syndrome-related protein 2 | | Authors: | Amaya, M.F, Dong, A, Adams-Cioaba, M.A, Guo, Y, MacKenzie, F, Kozieradzki, I, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-29 | | Release date: | 2009-06-16 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Studies of the Tandem Tudor Domains of Fragile X Mental Retardation Related Proteins FXR1 and FXR2.
Plos One, 5, 2010
|
|
3H9H
 
 | |
3HBX
 
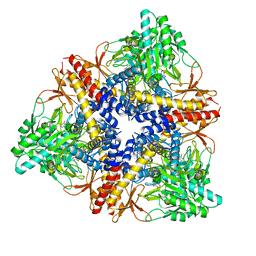 | | Crystal structure of GAD1 from Arabidopsis thaliana | | Descriptor: | Glutamate decarboxylase 1 | | Authors: | Gut, H, Dominici, P, Pilati, S, Gruetter, M.G, Capitani, G. | | Deposit date: | 2009-05-05 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.672 Å) | | Cite: | A common structural basis for pH- and calmodulin-mediated regulation in plant glutamate decarboxylase.
J.Mol.Biol., 392, 2009
|
|
3H9Z
 
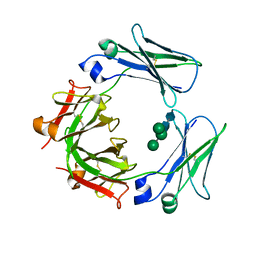 | | Crystal structure of the IgE-Fc3-4 domains | | Descriptor: | AMMONIUM ION, Ig epsilon chain C region, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Wurzburg, B.A. | | Deposit date: | 2009-04-30 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Conformational flexibility in immunoglobulin E-Fc 3-4 revealed in multiple crystal forms.
J.Mol.Biol., 393, 2009
|
|
3H4O
 
 | |
3H8N
 
 | | Crystal Structure Analysis of KIR2DS4 | | Descriptor: | Killer cell immunoglobulin-like receptor 2DS4 | | Authors: | Graef, T, Bushnell, D.A, Parham, P. | | Deposit date: | 2009-04-29 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A*11 while diminishing avidity for HLA-C.
J.Exp.Med., 206, 2009
|
|
3HC7
 
 | |
3H9Q
 
 | | Crystal structure of E. coli MccB + SeMet MccA | | Descriptor: | MccB protein, Microcin C7 ANALOG, SULFATE ION, ... | | Authors: | Regni, C.A, Roush, R.F, Miller, D, Nourse, A, Walsh, C.T, Schulman, B.A. | | Deposit date: | 2009-04-30 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic.
Embo J., 28, 2009
|
|
3H9F
 
 | | Crystal Structure of Human Dual Specificity Protein Kinase (TTK) in complex with a pyrimido-diazepin ligand | | Descriptor: | 9-cyclopentyl-2-(4-(4-hydroxypiperidin-1-yl)-2-methoxyphenylamino)-5-methyl-8,9-dihydro-5H-pyrimido[4,5-b][1,4]diazepin -6(7H)-one, Dual specificity protein kinase TTK, MAGNESIUM ION | | Authors: | Filippakopoulos, P, Soundararajan, M, Keates, T, Elkins, J.M, King, O, Fedorov, O, Picaud, S.S, Pike, A.C.W, Yue, W, Chaikuad, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Bountra, C, Kwiatkowski, N, Gray, N.S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-30 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Small-molecule kinase inhibitors provide insight into Mps1 cell cycle function.
Nat.Chem.Biol., 6, 2010
|
|
