4L6D
 
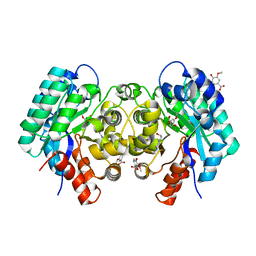 | | Crystal structure of 5-carboxyvanillate decarboxylase from Sphingomonas paucimobilis complexed with vanillic acid | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-HYDROXY-3-METHOXYBENZOATE, 5-carboxyvanillate decarboxylase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Vladimirova, A, Raushel, F.M, Almo, S.C. | | Deposit date: | 2013-06-12 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Crystal structure of 5-carboxyvanillate decarboxylase from Sphingomonas paucimobilis complexed with vanillic acid
To be Published
|
|
7R7O
 
 | |
7RRE
 
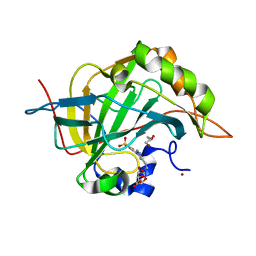 | | Carbonic Anhydrase II in complex with Beta-Galactose-2C | | Descriptor: | 2-[1-(1,1,3-trioxo-2,3-dihydro-1H-1lambda~6~,2-benzothiazol-6-yl)-1H-1,2,3-triazol-4-yl]ethyl beta-L-gulopyranoside, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | McKenna, R, Combs, J.E. | | Deposit date: | 2021-08-09 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Inhibition of Carbonic Anhydrase IX and Monocarboxylate Transporters 1 and 4 in breast cancer via novel inhibition with Beta-Galactose 2C
To Be Published
|
|
5AE8
 
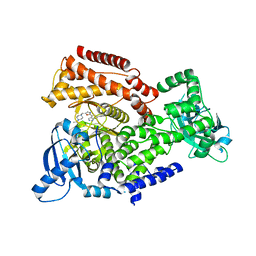 | | Crystal structure of mouse PI3 kinase delta in complex with GSK2269557 | | Descriptor: | 6-(1H-Indol-4-yl)-4-(5-{[4-(1-methylethyl)-1-piperazinyl]methyl}-1,3-oxazol-2-yl)-1H-indazole, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Down, K.D, Amour, A, Baldwin, I.R, Cooper, A.W.J, Deakin, A.M, Felton, L.M, Guntrip, S.B, Hardy, C, Harrison, Z.A, Jones, K.L, Jones, P, Keeling, S.E, Le, J, Livia, S, Lucas, F, Lunniss, C.J, Parr, N.J, Robinson, E, Rowland, P, Smith, S, Thomas, D.A, Vitulli, G, Washio, Y, Hamblin, N. | | Deposit date: | 2015-08-26 | | Release date: | 2015-09-16 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Optimization of Novel Indazoles as Highly Potent and Selective Inhibitors of Phosphoinositide 3-Kinase Gamma for the Treatment of Respiratory Disease.
J.Med.Chem., 58, 2015
|
|
5AE9
 
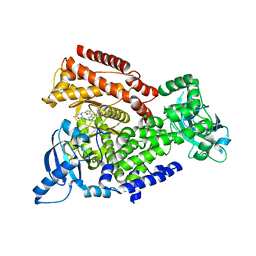 | | Crystal structure of mouse PI3 kinase delta in complex with GSK2292767 | | Descriptor: | N-[5-[4-(5-{[(2R,6S)-2,6-DIMETHYL-4-MORPHOLINYL]METHYL}-1,3-OXAZOL-2-YL)-1H-INDAZOL-6-YL]-2-(METHYLOXY)-3-PYRIDINYL]METHANESULFONAMIDE, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Down, K.D, Amour, A, Baldwin, I.R, Cooper, A.W.J, Deakin, A.M, Felton, L.M, Guntrip, S.B, Hardy, C, Harrison, Z.A, Jones, K.L, Jones, P, Keeling, S.E, Le, J, Livia, S, Lucas, F, Lunniss, C.J, Parr, N.J, Robinson, E, Rowland, P, Smith, S, Thomas, D.A, Vitulli, G, Washio, Y, Hamblin, N. | | Deposit date: | 2015-08-26 | | Release date: | 2015-09-16 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Optimization of Novel Indazoles as Highly Potent and Selective Inhibitors of Phosphoinositide 3-Kinase Gamma for the Treatment of Respiratory Disease.
J.Med.Chem., 58, 2015
|
|
8EHR
 
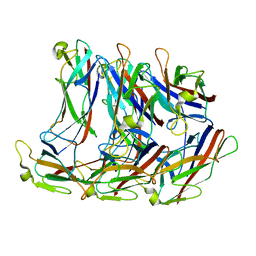 | |
6UUD
 
 | | Crystal structure of antibody 5D5 in complex with PfCSP N-terminal peptide | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 5D5 Antibody Fab, ... | | Authors: | Thai, E, Scally, S.W, Julien, J.P. | | Deposit date: | 2019-10-30 | | Release date: | 2020-07-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A high-affinity antibody against the CSP N-terminal domain lacks Plasmodium falciparum inhibitory activity.
J.Exp.Med., 217, 2020
|
|
8EHT
 
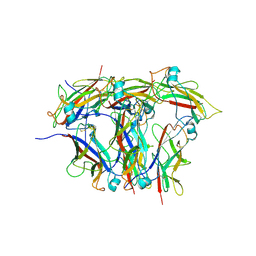 | |
2XWT
 
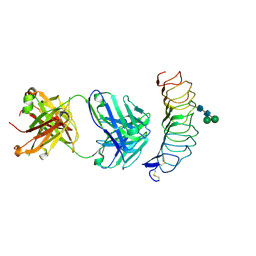 | | CRYSTAL STRUCTURE OF THE TSH RECEPTOR IN COMPLEX WITH A BLOCKING TYPE TSHR AUTOANTIBODY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, THYROID BLOCKING HUMAN AUTOANTIBODY K1-70 HEAVY CHAIN, ... | | Authors: | Sanders, J, Sanders, P, Young, S, Kabelis, K, Baker, S, Sullivan, A, Evans, M, Clark, J, Wilmot, J, Hu, X, Roberts, E, Powell, M, Nunez Miguel, R, Furmaniak, J, Rees Smith, B. | | Deposit date: | 2010-11-05 | | Release date: | 2011-03-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Tsh Receptor (Tshr) Bound to a Blocking-Type Tshr Autoantibody.
J.Mol.Endocrinol., 46, 2011
|
|
7R1U
 
 | | Crystal structure of SARS-CoV-2 nsp10/nsp16 in complex with the WZ16 inhibitor | | Descriptor: | (2S,5S)-2,6-diamino-5-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}hexanoic acid, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Klima, M, Boura, E, Li, F, Yazdi, A.K, Vedadi, M. | | Deposit date: | 2022-02-03 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10-nsp16 in complex with small molecule inhibitors, SS148 and WZ16.
Protein Sci., 31, 2022
|
|
1A3B
 
 | | COMPLEX OF HUMAN ALPHA-THROMBIN WITH THE BIFUNCTIONAL BORONATE INHIBITOR BOROLOG1 | | Descriptor: | ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), Hirudin, ... | | Authors: | Skordalakes, E, Elgendy, S, Dodson, G, Goodwin, C.A, Green, D, Scully, M.F, Freyssinet, J.H, Kakkar, V.V, Deadman, J. | | Deposit date: | 1998-01-20 | | Release date: | 1998-06-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bifunctional peptide boronate inhibitors of thrombin: crystallographic analysis of inhibition enhanced by linkage to an exosite 1 binding peptide.
Biochemistry, 37, 1998
|
|
7R1T
 
 | | Crystal structure of SARS-CoV-2 nsp10/nsp16 in complex with the SS148 inhibitor | | Descriptor: | (2~{S})-2-azanyl-4-[[(2~{S},3~{S},4~{R},5~{R})-5-(4-azanyl-5-cyano-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methylsulfanyl]butanoic acid, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Klima, M, Boura, E, Li, F, Yazdi, A.K, Vedadi, M. | | Deposit date: | 2022-02-03 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10-nsp16 in complex with small molecule inhibitors, SS148 and WZ16.
Protein Sci., 31, 2022
|
|
2VZ5
 
 | | Structure of the PDZ domain of Tax1 (human T-cell leukemia virus type I) binding protein 3 | | Descriptor: | CHLORIDE ION, IMIDAZOLE, TAX1-BINDING PROTEIN 3, ... | | Authors: | Murray, J.W, Shafqat, N, Yue, W, Pilka, E, Johannsson, C, Salah, E, Cooper, C, Elkins, J.M, Pike, A.C, Roos, A, Filippakopoulos, P, von Delft, F, Wickstroem, M, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Oppermann, U. | | Deposit date: | 2008-07-30 | | Release date: | 2008-08-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.738 Å) | | Cite: | The Structure of the Pdz Domain of Tax1BP
To be Published
|
|
6EXF
 
 | | Crystal structure of the complex Fe(II)/alpha-ketoglutarate dependent dioxygenase KDO5 with Fe(II)/Lysine | | Descriptor: | FE (III) ION, GLYCEROL, L-lysine 4-hydroxylase, ... | | Authors: | Isabet, T, Stura, E, Legrand, P, Zaparucha, A, Bastard, K. | | Deposit date: | 2017-11-08 | | Release date: | 2018-11-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Studies based on two Lysine Dioxygenases with Distinct Regioselectivity Brings Insights Into Enzyme Specificity within the Clavaminate Synthase-Like Family.
Sci Rep, 8, 2018
|
|
7RRF
 
 | | Carbonic Anhydrase IX-mimic in complex with Beta-Galactose_2C | | Descriptor: | 2-[1-(1,1,3-trioxo-2,3-dihydro-1H-1lambda~6~,2-benzothiazol-6-yl)-1H-1,2,3-triazol-4-yl]ethyl beta-L-gulopyranoside, Carbonic anhydrase 2, ZINC ION | | Authors: | Combs, J.E, McKenna, R. | | Deposit date: | 2021-08-09 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Inhibition of Carbonic Anhydrase IX and Monocarboxylate Transporters 1 and 4 in breast cancer via novel inhibition with Beta-Galactose 2C
To Be Published
|
|
8CPD
 
 | | Cryo-EM structure of CRaf dimer with 14:3:3 | | Descriptor: | 14-3-3 protein zeta isoform X1, RAF proto-oncogene serine/threonine-protein kinase | | Authors: | Dedden, D, Graedler, U, Schwarz, D, Thomsen, M, Leuthner, B, Schneider, E, Nitsche, J. | | Deposit date: | 2023-03-02 | | Release date: | 2024-02-21 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Cryo-EM Structures of CRAF 2 /14-3-3 2 and CRAF 2 /14-3-3 2 /MEK1 2 Complexes.
J.Mol.Biol., 436, 2024
|
|
5VA7
 
 | | Glucocorticoid Receptor DNA Binding Domain - IL11 AP-1 recognition element Complex | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*TP*GP*AP*GP*TP*CP*AP*GP*GP*AP*TP*G)-3'), DNA (5'-D(*CP*AP*TP*CP*CP*TP*GP*AP*CP*TP*CP*AP*CP*CP*CP*T)-3'), Glucocorticoid receptor, ... | | Authors: | Weikum, E.R, Ortlund, E.A. | | Deposit date: | 2017-03-24 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | Tethering not required: the glucocorticoid receptor binds directly to activator protein-1 recognition motifs to repress inflammatory genes.
Nucleic Acids Res., 45, 2017
|
|
4H2E
 
 | | Crystal structure of an MMP twin inhibitor complexing two MMP-9 catalytic domains | | Descriptor: | ACETATE ION, BICINE, CALCIUM ION, ... | | Authors: | Stura, E.A, Vera, L, Cassar-Lajeunesse, E, Nuti, E, Catalani, M.P, Dive, V, Rossello, A. | | Deposit date: | 2012-09-12 | | Release date: | 2013-04-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Crystallization of bi-functional ligand protein complexes.
J.Struct.Biol., 182, 2013
|
|
6F2W
 
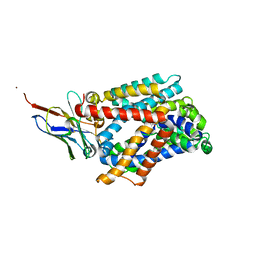 | | Bacterial asc transporter crystal structure in open to in conformation | | Descriptor: | ALPHA-AMINOISOBUTYRIC ACID, Nanobody 74, Putative amino acid/polyamine transport protein, ... | | Authors: | Fort, J, Errasti-Murugarren, E, Carpena, X, Palacin, M, Fita, I. | | Deposit date: | 2017-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | L amino acid transporter structure and molecular bases for the asymmetry of substrate interaction.
Nat Commun, 10, 2019
|
|
3KJV
 
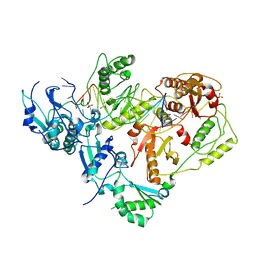 | | HIV-1 reverse transcriptase in complex with DNA | | Descriptor: | 5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*(DOC))-3', 5'-D(*AP*TP*GP*GP*TP*GP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3', MAGNESIUM ION, ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2009-11-03 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Visualizing the molecular interactions of a nucleotide analog, GS-9148, with HIV-1 reverse transcriptase-DNA complex.
J.Mol.Biol., 397, 2010
|
|
5UMK
 
 | |
198L
 
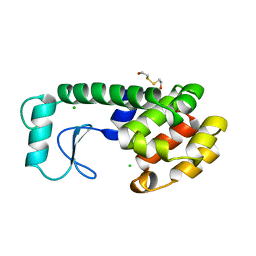 | | THERMODYNAMIC AND STRUCTURAL COMPENSATION IN "SIZE-SWITCH" CORE-REPACKING VARIANTS OF T4 LYSOZYME | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, LYSOZYME | | Authors: | Baldwin, E, Xu, J, Hajiseyedjavadi, O, Matthews, B.W. | | Deposit date: | 1995-11-06 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Thermodynamic and structural compensation in "size-switch" core repacking variants of bacteriophage T4 lysozyme.
J.Mol.Biol., 259, 1996
|
|
8C7D
 
 | | Structure of the GEF Kalirin DH2 Domain | | Descriptor: | Kalirin | | Authors: | Callens, M.C, Thompson, A.P, Gray, J.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-01-14 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure-based alignment and analysis of RHO selectivity of RHO-DBL GTPase exchange factors
to be published
|
|
5F7N
 
 | | Blood group antigen binding adhesin BabA of Helicobacter pylori strain 17875 in complex with blood group A Lewis b pentasaccharide | | Descriptor: | Adhesin binding fucosylated histo-blood group antigen, Nanobody Nb-ER19, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Moonens, K, Gideonsson, P, Subedi, S, Romao, E, Oscarson, S, Muyldermans, S, Boren, T, Remaut, H. | | Deposit date: | 2015-12-08 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural Insights into Polymorphic ABO Glycan Binding by Helicobacter pylori.
Cell Host Microbe, 19, 2016
|
|
8BYL
 
 | | Cryo-EM structure of SKP1-SKP2-CKS1 from the SCFSKP2 E3 ligase complex | | Descriptor: | Cyclin-dependent kinase inhibitor 1B, Cyclin-dependent kinases regulatory subunit 1, S-phase kinase-associated protein 1, ... | | Authors: | Rowland, R.J, Salamina, M, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2022-12-13 | | Release date: | 2023-06-28 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of SKP1-SKP2-CKS1 in complex with CDK2-cyclin A-p27KIP1.
Sci Rep, 13, 2023
|
|
