7AGM
 
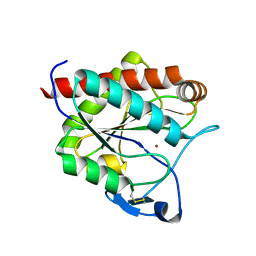 | | Crystal structure of the N-acetylmuramyl-L-alanine amidase, Ami1, from Mycobacterium smegmatis | | Descriptor: | N-acetylmuramoyl-L-alanine amidase, ZINC ION | | Authors: | Blaise, M, Alsarraf, M.A.B. | | Deposit date: | 2020-09-23 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Functional Characterization of the N -Acetylmuramyl-l-Alanine Amidase, Ami1, from Mycobacterium abscessus .
Cells, 9, 2020
|
|
7A1D
 
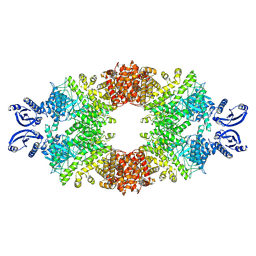 | | Cryo-EM map of the large glutamate dehydrogenase composed of 180 kDa subunits from Mycobacterium smegmatis (open conformation) | | Descriptor: | NAD-specific glutamate dehydrogenase | | Authors: | Lazaro, M, Melero, R, Huet, C, Lopez-Alonso, J.P, Delgado, S, Dodu, A, Bruch, E.M, Abriata, L.A, Alzari, P.M, Valle, M, Lisa, M.N. | | Deposit date: | 2020-08-12 | | Release date: | 2021-06-09 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.19 Å) | | Cite: | 3D architecture and structural flexibility revealed in the subfamily of large glutamate dehydrogenases by a mycobacterial enzyme.
Commun Biol, 4, 2021
|
|
6OR2
 
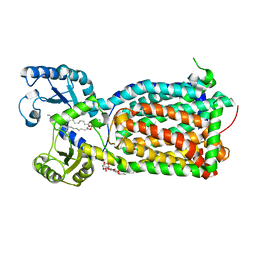 | | MmpL3 is a lipid transporter that binds trehalose monomycolate and phosphatidylethanolamine | | Descriptor: | (1S)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, DODECYL-BETA-D-MALTOSIDE, Membrane protein, ... | | Authors: | Su, C.-C. | | Deposit date: | 2019-04-29 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | MmpL3 is a lipid transporter that binds trehalose monomycolate and phosphatidylethanolamine.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5O61
 
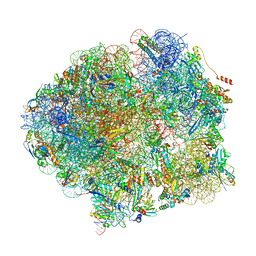 | | The complete structure of the Mycobacterium smegmatis 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Hentschel, J, Burnside, C, Mignot, I, Leibundgut, M, Boehringer, D, Ban, N. | | Deposit date: | 2017-06-03 | | Release date: | 2017-07-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | The Complete Structure of the Mycobacterium smegmatis 70S Ribosome.
Cell Rep, 20, 2017
|
|
6DZK
 
 | | Cryo-EM Structure of Mycobacterium smegmatis C(minus) 30S ribosomal subunit with MPY | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Sharma, M.R, Li, Y, Korripella, R, Yang, Y, Kaushal, P.S, Lin, Q, Wade, J.T, Gray, A.G, Derbyshire, K.M, Agrawal, R.K, Ojha, A. | | Deposit date: | 2018-07-05 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Zinc depletion induces ribosome hibernation in mycobacteria.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7Q59
 
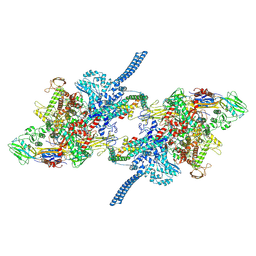 | |
6DZI
 
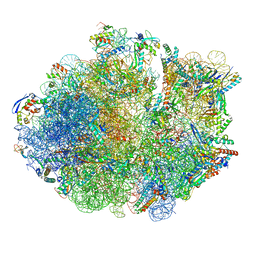 | | Cryo-EM Structure of Mycobacterium smegmatis 70S C(minus) ribosome 70S-MPY complex | | Descriptor: | 16S rRNA, 23 S rRNA (3119-MER), 30S ribosomal protein S10, ... | | Authors: | Sharma, M.R, Li, Y, Korripella, R, Yang, Y, Kaushal, P.S, Lin, Q, Wade, J.T, Gray, A.G, Derbyshire, K.M, Agrawal, R.K, Ojha, A. | | Deposit date: | 2018-07-05 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Zinc depletion induces ribosome hibernation in mycobacteria.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7PP4
 
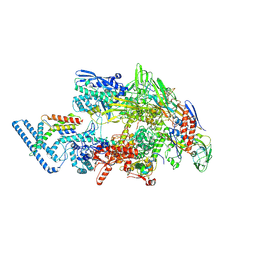 | |
7O4V
 
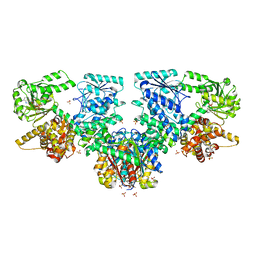 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in complex with oxidized nicotinamide adenine dinucleotide | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative acyltransferase Rv0859, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2021-04-07 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Substrate specificity and conformational flexibility properties of the Mycobacterium tuberculosis beta-oxidation trifunctional enzyme.
J.Struct.Biol., 213, 2021
|
|
7O4Q
 
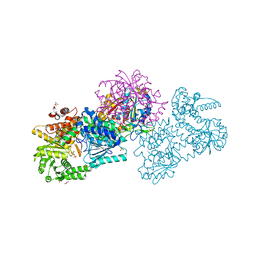 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme in space group C2221 (unliganded) | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, GLYCEROL, Putative acyltransferase Rv0859, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2021-04-07 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate specificity and conformational flexibility properties of the Mycobacterium tuberculosis beta-oxidation trifunctional enzyme.
J.Struct.Biol., 213, 2021
|
|
7O4R
 
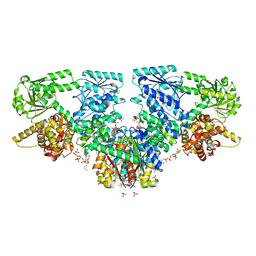 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme with Coenzyme A bound at the thiolase active sites and additional binding site (CoA(HAD/KAT)) | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, COENZYME A, GLYCEROL, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2021-04-07 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Substrate specificity and conformational flexibility properties of the Mycobacterium tuberculosis beta-oxidation trifunctional enzyme.
J.Struct.Biol., 213, 2021
|
|
7O4T
 
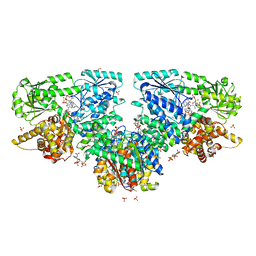 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme with Coenzyme A bound at the hydratase, thiolase active sites and possible additional binding site (CoA(ECH/HAD)) | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-DIPHOSPHATE, 3-hydroxyacyl-CoA dehydrogenase, COENZYME A, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2021-04-07 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate specificity and conformational flexibility properties of the Mycobacterium tuberculosis beta-oxidation trifunctional enzyme.
J.Struct.Biol., 213, 2021
|
|
7O4S
 
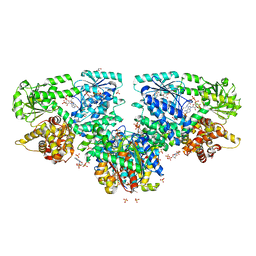 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme with Coenzyme A bound at the hydratase, thiolase active sites and additional binding site (CoA(ECH2)) | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-DIPHOSPHATE, 3-hydroxyacyl-CoA dehydrogenase, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2021-04-07 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Substrate specificity and conformational flexibility properties of the Mycobacterium tuberculosis beta-oxidation trifunctional enzyme.
J.Struct.Biol., 213, 2021
|
|
7REF
 
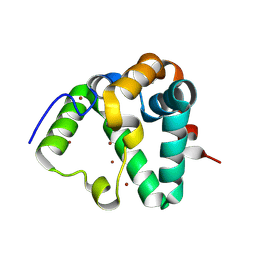 | | Structure of MS3494 from Mycobacterium smegmatis | | Descriptor: | BROMIDE ION, MS3494 | | Authors: | Kent, J.E, Aleshin, A.E, Zhang, L, Niederweis, M, Marassi, F.M. | | Deposit date: | 2021-07-12 | | Release date: | 2021-08-18 | | Last modified: | 2022-06-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A periplasmic cinched protein is required for siderophore secretion and virulence of Mycobacterium tuberculosis.
Nat Commun, 13, 2022
|
|
6ZT4
 
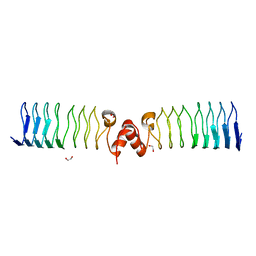 | | Pentapeptide repeat protein MfpA from Mycobacterium smegmatis | | Descriptor: | 1,2-ETHANEDIOL, Pentapeptide repeat protein MfpA | | Authors: | Feng, L, Mundy, J.E.A, Stevenson, C.E.M, Mitchenall, L.A, Lawson, D.M, Mi, K, Maxwell, A. | | Deposit date: | 2020-07-17 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The pentapeptide-repeat protein, MfpA, interacts with mycobacterial DNA gyrase as a DNA T-segment mimic.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7JG7
 
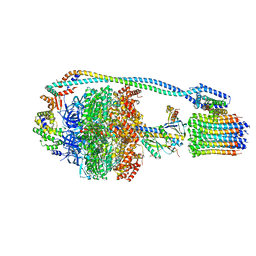 | | Cryo-EM structure of bedaquiline-free Mycobacterium smegmatis ATP synthase rotational state 3 (backbone model) | | Descriptor: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit a, ... | | Authors: | Guo, H, Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2020-07-18 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
7JG8
 
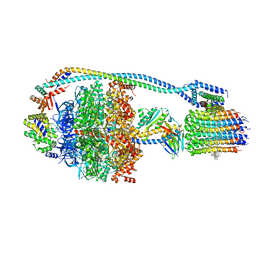 | | Cryo-EM structure of bedaquiline-saturated Mycobacterium smegmatis ATP synthase rotational state 1 (backbone model) | | Descriptor: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit a, ... | | Authors: | Guo, H, Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2020-07-18 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
7JGA
 
 | | Cryo-EM structure of bedaquiline-saturated Mycobacterium smegmatis ATP synthase rotational state 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Guo, H, Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2020-07-18 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
7JG6
 
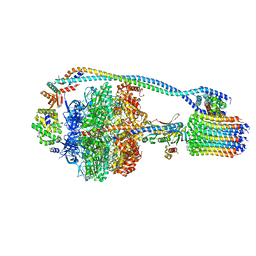 | | Cryo-EM structure of bedaquiline-free Mycobacterium smegmatis ATP synthase rotational state 2 (backbone model) | | Descriptor: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit a, ... | | Authors: | Guo, H, Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2020-07-18 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
7JG5
 
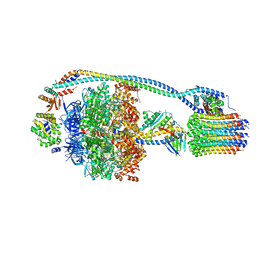 | | Cryo-EM structure of bedaquiline-free Mycobacterium smegmatis ATP synthase rotational state 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Guo, H, Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2020-07-18 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
7JG9
 
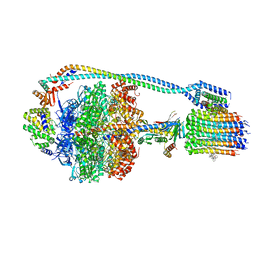 | | Cryo-EM structure of bedaquiline-saturated mycobacterium smegmatis ATP synthase rotational state 2 (backbone model) | | Descriptor: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit a, ... | | Authors: | Guo, H, Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2020-07-18 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
7O1I
 
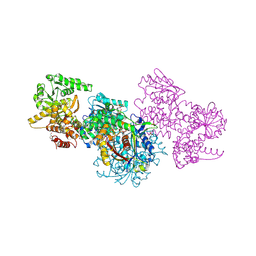 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme alpha-E141A mutant | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, COENZYME A, GLYCEROL, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate specificity and conformational flexibility properties of the Mycobacterium tuberculosis beta-oxidation trifunctional enzyme.
J.Struct.Biol., 213, 2021
|
|
7O1G
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme alpha-E141A-H462A, beta-C92A mutant | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, Putative acyltransferase Rv0859, SULFATE ION | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Substrate specificity and conformational flexibility properties of the Mycobacterium tuberculosis beta-oxidation trifunctional enzyme.
J.Struct.Biol., 213, 2021
|
|
7O1K
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme alpha-E141A, beta-C92A mutant | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, GLYCEROL, Putative acyltransferase Rv0859, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Substrate specificity and conformational flexibility properties of the Mycobacterium tuberculosis beta-oxidation trifunctional enzyme.
J.Struct.Biol., 213, 2021
|
|
7O1L
 
 | | Structure of Mycobacterium tuberculosis beta-oxidation trifunctional enzyme alpha-H462A mutant | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, COENZYME A, GLYCEROL, ... | | Authors: | Dalwani, S, Wierenga, R.K, Venkatesan, R. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Substrate specificity and conformational flexibility properties of the Mycobacterium tuberculosis beta-oxidation trifunctional enzyme.
J.Struct.Biol., 213, 2021
|
|
