4GDP
 
 | | Yeast polyamine oxidase FMS1, N195A mutant | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Polyamine oxidase FMS1, TETRAETHYLENE GLYCOL | | Authors: | Taylor, A.B, Adachi, M.S, Hart, P.J, Fitzpatrick, P.F. | | Deposit date: | 2012-08-01 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9998 Å) | | Cite: | Mechanistic and Structural Analyses of the Roles of Active Site Residues in Yeast Polyamine Oxidase Fms1: Characterization of the N195A and D94N Enzymes.
Biochemistry, 51, 2012
|
|
3VY9
 
 | | Crystal structure of PorB from Neisseria meningitidis in complex with cesium ion, space group H32 | | Descriptor: | CESIUM ION, Outer membrane protein | | Authors: | Kattner, C, Zaucha, J, Jaenecke, F, Zachariae, U, Tanabe, M. | | Deposit date: | 2012-09-21 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Identification of a cation transport pathway in Neisseria meningitidis PorB.
Proteins, 81, 2013
|
|
4GED
 
 | | Crystal Structure of the Leishmania Major Peroxidase-Cytochrome C Complex | | Descriptor: | Ascorbate peroxidase, Cytochrome c, HEME C, ... | | Authors: | Jasion, V.S, Doukov, T, Pineda, S.H, Li, H, Poulos, T.L. | | Deposit date: | 2012-08-01 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of the Leishmania major peroxidase-cytochrome c complex.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2TRC
 
 | | PHOSDUCIN/TRANSDUCIN BETA-GAMMA COMPLEX | | Descriptor: | GADOLINIUM ATOM, PHOSDUCIN, TRANSDUCIN | | Authors: | Gaudet, R, Bohm, A, Sigler, P.B. | | Deposit date: | 1997-01-06 | | Release date: | 1997-06-05 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure at 2.4 angstroms resolution of the complex of transducin betagamma and its regulator, phosducin.
Cell(Cambridge,Mass.), 87, 1996
|
|
4GI1
 
 | | Structure of the complex of three phase partition treated lipase from Thermomyces lanuginosa with 16-hydroxypalmitic acid at 2.4 A resolution | | Descriptor: | 16-hydroxyhexadecanoic acid, GLYCEROL, Lipase | | Authors: | Kumar, M, Sinha, M, Mukherjee, J, Gupta, M.N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2012-08-08 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structure of the complex of three phase partition treated lipase from Thermomyces lanuginosa with 16-hydroxypalmitic acid at 2.4 A resolution
To be published
|
|
3W1N
 
 | | Crystal structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with 5-iodoorotate | | Descriptor: | 5-iodo-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, COBALT HEXAMMINE(III), Dihydroorotate dehydrogenase (fumarate), ... | | Authors: | Inaoka, D.K, Iida, M, Tabuchi, T, Lee, N, Matsuoka, S, Shiba, T, Sakamoto, K, Suzuki, S, Balogun, E.O, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2012-11-19 | | Release date: | 2013-11-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with 5-iodoorotate
To be Published
|
|
3VY5
 
 | |
3W2C
 
 | |
4GGR
 
 | | The structure of apo bradavidin2 (Form A) | | Descriptor: | Bradavidin 2 | | Authors: | Livnah, O, Meir, A. | | Deposit date: | 2012-08-07 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The highly dynamic oligomeric structure of bradavidin II is unique among avidin proteins.
Protein Sci., 22, 2013
|
|
4GH0
 
 | | Crystal structure of D48V mutant of human GLTP bound with 12:0 monosulfatide | | Descriptor: | Glycolipid transfer protein, N-{(2S,3R,4E)-3-hydroxy-1-[(3-O-sulfo-beta-D-galactopyranosyl)oxy]octadec-4-en-2-yl}dodecanamide, PENTADECANE | | Authors: | Samygina, V.R, Cabo-Bilbao, A, Ochoa-Lizarralde, B, Popov, A.N, Malinina, L. | | Deposit date: | 2012-08-07 | | Release date: | 2013-04-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural insights into lipid-dependent reversible dimerization of human GLTP.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2RHF
 
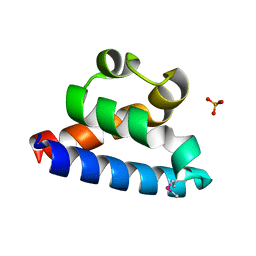 | | D. radiodurans RecQ HRDC domain 3 | | Descriptor: | DNA helicase RecQ, PHOSPHATE ION | | Authors: | Keck, J.L, Killoran, M.P. | | Deposit date: | 2007-10-09 | | Release date: | 2008-04-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure and function of the regulatory C-terminal HRDC domain from Deinococcus radiodurans RecQ.
Nucleic Acids Res., 36, 2008
|
|
3VDG
 
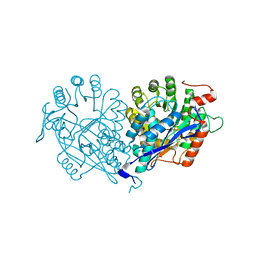 | | Crystal structure of enolase MSMEG_6132 (TARGET EFI-502282) from Mycobacterium smegmatis str. MC2 155 complexed with formate and acetate | | Descriptor: | ACETATE ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-05 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of enolase MSMEG_6132 FROM Mycobacterium smegmatis
To be Published
|
|
4ZY6
 
 | |
5OLL
 
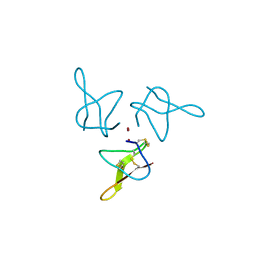 | | Crystal structure of gurmarin, a sweet taste suppressing polypeptide | | Descriptor: | Gurmarin, NICKEL (II) ION | | Authors: | Sigoillot, M, Neiers, F, Legrand, P, Roblin, P, Briand, L. | | Deposit date: | 2017-07-28 | | Release date: | 2018-08-08 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Crystal Structure of Gurmarin, a Sweet Taste-Suppressing Protein: Identification of the Amino Acid Residues Essential for Inhibition.
Chem. Senses, 43, 2018
|
|
3V9S
 
 | |
3VFH
 
 | |
5A3X
 
 | | DYRK1A in complex with hydroxy benzothiazole fragment | | Descriptor: | DUAL SPECIFICITY TYROSINE-PHOSPHORYLATION-REGULATED KINASE 1A, N-(5-oxidanyl-1,3-benzothiazol-2-yl)ethanamide | | Authors: | Rothweiler, U. | | Deposit date: | 2015-06-03 | | Release date: | 2016-06-29 | | Last modified: | 2019-02-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Probing the ATP-Binding Pocket of Protein Kinase Dyrk1A with Benzothiazole Fragment Molecules
J.Med.Chem., 59, 2016
|
|
4WI8
 
 | | Structural mapping of the human IgG1 binding site for FcRn: hu3S193 Fc mutation Y436A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose, ... | | Authors: | Farrugia, W, Burvenich, I.J.G, Scott, A.M, Ramsland, P.A. | | Deposit date: | 2014-09-25 | | Release date: | 2015-09-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional mapping of human IgG1 binding site for FcRn in vivo using human FcRn transgenic mice
To Be Published
|
|
4GJR
 
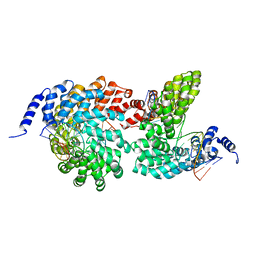 | | Crystal structure of the TAL effector dHax3 bound to methylated dsDNA | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*AP*GP*GP*TP*AP*GP*AP*GP*GP*GP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*(5CM)P*TP*AP*(5CM)P*CP*TP*CP*(5CM)P*CP*T)-3'), Hax3 | | Authors: | Yan, N, Deng, D, Yan, C.Y, Yin, P, Pan, X.J, Shi, Y.G. | | Deposit date: | 2012-08-10 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of a protein complex
To be Published
|
|
4GG4
 
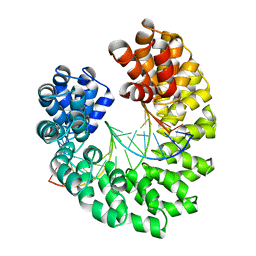 | | Crystal structure of the TAL effector dHax3 bound to specific DNA-RNA hybrid | | Descriptor: | DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*TP*AP*TP*CP*TP*CP*TP*CP*T)-3'), Hax3, RNA (5'-R(*AP*GP*AP*GP*AP*GP*AP*UP*AP*AP*AP*GP*GP*GP*AP*CP*A)-3') | | Authors: | Yin, P, Deng, D, Yan, C.Y, Pan, X.J, Yan, N, Shi, Y.G. | | Deposit date: | 2012-08-05 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Specific DNA-RNA hybrid recognition by TAL effectors
Cell Rep, 2, 2012
|
|
2TGD
 
 | |
4GGK
 
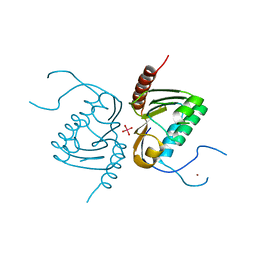 | | Crystal structure of Zucchini from mouse (mZuc / PLD6 / MitoPLD) bound to tungstate | | Descriptor: | Mitochondrial cardiolipin hydrolase, TUNGSTATE(VI)ION, ZINC ION | | Authors: | Ipsaro, J.J, Haase, A.D, Hannon, G.J, Joshua-Tor, L. | | Deposit date: | 2012-08-06 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structural biochemistry of Zucchini implicates it as a nuclease in piRNA biogenesis.
Nature, 491, 2012
|
|
3VDA
 
 | | E. coli (lacZ) beta-galactosidase (N460T) | | Descriptor: | Beta-galactosidase, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Wheatley, R.W, Kappelhoff, J.C, Hahn, J.N, Dugdale, M.L, Dutkoski, M.J, Tamman, S.D, Fraser, M.E, Huber, R.E. | | Deposit date: | 2012-01-04 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substitution for Asn460 cripples {beta}-galactosidase (Escherichia coli) by increasing substrate affinity and decreasing transition state stability.
Arch.Biochem.Biophys., 521, 2012
|
|
4GGT
 
 | | Structure of apo Bradavidin2 (Form B) | | Descriptor: | Bradavidin 2 | | Authors: | Livnah, O, Meir, A. | | Deposit date: | 2012-08-07 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.693 Å) | | Cite: | The highly dynamic oligomeric structure of bradavidin II is unique among avidin proteins.
Protein Sci., 22, 2013
|
|
2SHK
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF SHIKIMATE KINASE FROM ERWINIA CHRYSANTHEMI COMPLEXED WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SHIKIMATE KINASE | | Authors: | Krell, T, Coggins, J.R, Lapthorn, A.J. | | Deposit date: | 1997-10-27 | | Release date: | 1998-11-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallization and preliminary X-ray crystallographic analysis of shikimate kinase from Erwinia chrysanthemi.
Acta Crystallogr.,Sect.D, 53, 1997
|
|
