2XXH
 
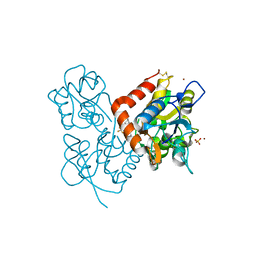 | | Crystal structure of 1-(4-(2-oxo-2-(1-pyrrolidinyl)ethyl)phenyl)-3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazole in complex with the ligand binding domain of the Rat GluA2 receptor and glutamate at 1.5A resolution. | | Descriptor: | 1-{4-[2-OXO-2-(1-PYRROLIDINYL)ETHYL]PHENYL}-3-( TRIFLUOROMETHYL)-4,5,6,7-TETRAHYDRO-1H-INDAZOLE, GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, ... | | Authors: | Ward, S.E, Harries, M, Aldegheri, L, Austin, N.E, Ballantine, S, Ballini, E, Bradley, D.M, Bax, B.D, Clarke, B.P, Harris, A.J, Harrison, S.A, Melarange, R.A, Mookherjee, C, Mosley, J, DalNegro, G, Oliosi, B, Smith, K.J, Thewlis, K.M, Woollard, P.M, Yusaf, S.P. | | Deposit date: | 2010-11-10 | | Release date: | 2011-04-06 | | Last modified: | 2011-09-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Integration of Lead Optimization with Crystallography for a Membrane-Bound Ion Channel Target: Discovery of a New Class of Ampa Receptor Positive Allosteric Modulators.
J.Med.Chem., 54, 2011
|
|
2XX8
 
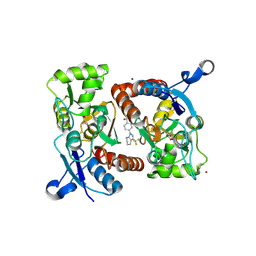 | | Crystal structure of N,N-dimethyl-4-(3-(trifluoromethyl)-4,5,6,7- tetrahydro-1H-indazol-1-yl)benzamide in complex with the ligand binding domain of the Rat GluA2 receptor and glutamate at 2.2A resolution. | | Descriptor: | GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, N,N-DIMETHYL-4-[3-(TRIFLUOROMETHYL)-4,5,6,7-TETRAHYDRO-1H-INDAZOL-1-YL]BENZAMIDE, ... | | Authors: | Ward, S.E, Harries, M, Aldegheri, L, Austin, N.E, Ballantine, S, Ballini, E, Bradley, D.M, Bax, B.D, Clarke, B.P, Harris, A.J, Harrison, S.A, Melarange, R.A, Mookherjee, C, Mosley, J, DalNegro, G, Oliosi, B, Smith, K.J, Thewlis, K.M, Woollard, P.M, Yusaf, S.P. | | Deposit date: | 2010-11-09 | | Release date: | 2011-04-27 | | Last modified: | 2011-09-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Integration of Lead Optimization with Crystallography for a Membrane-Bound Ion Channel Target: Discovery of a New Class of Ampa Receptor Positive Allosteric Modulators.
J.Med.Chem., 54, 2011
|
|
6UXR
 
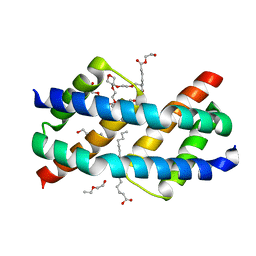 | | Crystal structure of BAK core domain BH3-groove-dimer in complex with LysoPC | | Descriptor: | Bcl-2 homologous antagonist/killer, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, ... | | Authors: | Cowan, A.D, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-11-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | BAK core dimers bind lipids and can be bridged by them.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4R9T
 
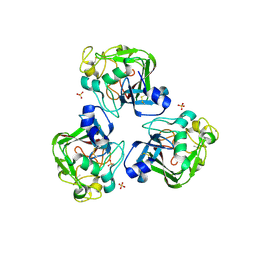 | | L-ficolin complexed to sulphates | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Laffly, E, Lacroix, M, Martin, L, Vassal-Stermann, E, Thielens, N, Gaboriaud, C. | | Deposit date: | 2014-09-08 | | Release date: | 2014-11-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Human ficolin-2 recognition versatility extended: An update on the binding of ficolin-2 to sulfated/phosphated carbohydrates.
Febs Lett., 588, 2014
|
|
7JVR
 
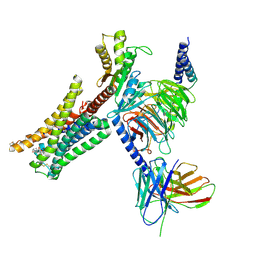 | | Cryo-EM structure of Bromocriptine-bound dopamine receptor 2 in complex with Gi protein | | Descriptor: | Antibody fragment ScFv16, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhuang, Y, Xu, P, Mao, C, Wang, L, Krumm, B, Zhou, X.E, Huang, S, Liu, H, Cheng, X, Huang, X.-P, Sheng, D.-D, Xu, T, Liu, Y.-F, Wang, Y, Guo, J, Jiang, Y, Jiang, H, Melcher, K, Roth, B.L, Zhang, Y, Zhang, C, Xu, H.E. | | Deposit date: | 2020-08-22 | | Release date: | 2021-02-24 | | Last modified: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into the human D1 and D2 dopamine receptor signaling complexes.
Cell, 184, 2021
|
|
4K0J
 
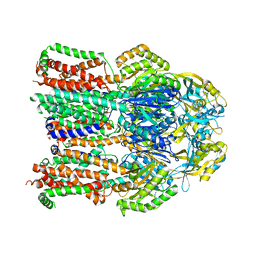 | | X-ray crystal structure of a heavy metal efflux pump, crystal form I | | Descriptor: | Heavy metal cation tricomponent efflux pump ZneA(CzcA-like), ZINC ION | | Authors: | Pak, J.E, Stroud, R.M, Ngonlong Ekende, E, Vandenbussche, G, Center for Structures of Membrane Proteins (CSMP) | | Deposit date: | 2013-04-04 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of intermediate transport states of ZneA, a Zn(II)/proton antiporter.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5LXI
 
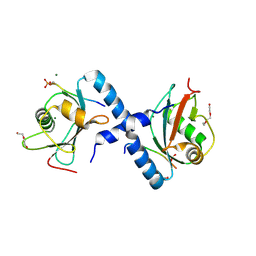 | | GABARAP-L1 ATG4B LIR Complex | | Descriptor: | 1,2-ETHANEDIOL, Cysteine protease ATG4B, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mouilleron, S, Skytte Rasmussen, M, Kumar Shrestha, B, Wirth, M, Bowitz Larsen, K, Abudu Princely, Y, Sjottem, E, Tooze, S, Lamark, T, Johansen, T, Lee, R. | | Deposit date: | 2016-09-21 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | ATG4B contains a C-terminal LIR motif important for binding and efficient cleavage of mammalian orthologs of yeast Atg8.
Autophagy, 13, 2017
|
|
5W3H
 
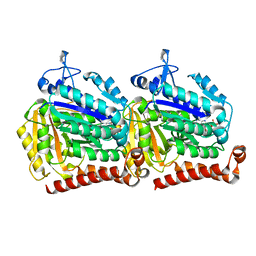 | | Yeast microtubule stabilized with epothilone | | Descriptor: | EPOTHILONE A, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Howes, S.C, Geyer, E.A, LaFrance, B, Zhang, R, Kellogg, E.H, Westermann, S, Rice, L.M, Nogales, E. | | Deposit date: | 2017-06-07 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural differences between yeast and mammalian microtubules revealed by cryo-EM.
J. Cell Biol., 216, 2017
|
|
2OHX
 
 | |
5B6M
 
 | |
8EEV
 
 | | Venezuelan equine encephalitis virus-like particle in complex with Fab SKT-20 | | Descriptor: | Coat protein, Fab SKT20 heavy chain, Fab SKT20 light chain | | Authors: | Tsybovsky, Y, Pletnev, S, Verardi, R, Roederer, M, Kwong, P.D. | | Deposit date: | 2022-09-07 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Vaccine elicitation and structural basis for antibody protection against alphaviruses.
Cell, 186, 2023
|
|
7U5O
 
 | | CRYSTAL STRUCTURE OF THE BONE MORPHOGENETIC PROTEIN RECEPTOR TYPE 2 LIGAND BINDING DOMAIN IN COMPLEX WITH ACTIVIN-B | | Descriptor: | Bone morphogenetic protein receptor type-2, Inhibin beta B chain | | Authors: | Chu, K.Y, Malik, A, Thamilselvan, V, Martinez-Hackert, E. | | Deposit date: | 2022-03-02 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Type II BMP and activin receptors BMPR2 and ACVR2A share a conserved mode of growth factor recognition.
J.Biol.Chem., 298, 2022
|
|
6Q7J
 
 | | GH3 exo-beta-xylosidase (XlnD) in complex with xylobiose aziridine activity based probe | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Davies, G.J, Rowland, R.J, Wu, L, Moroz, O, Blagova, E. | | Deposit date: | 2018-12-13 | | Release date: | 2019-06-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Dynamic and Functional Profiling of Xylan-Degrading Enzymes inAspergillusSecretomes Using Activity-Based Probes.
Acs Cent.Sci., 5, 2019
|
|
4R2Y
 
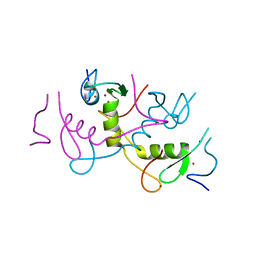 | | Crystal structure of APC11 RING domain | | Descriptor: | Anaphase-promoting complex subunit 11, ZINC ION | | Authors: | Brown, N.G, Watson, E.R, Weissmann, F, Jarvis, M.A, Vanderlinden, R, Grace, C.R.R, Frye, J.J, Dube, P, Qiao, R, Petzold, G, Cho, S.E, Alsharif, O, Bao, J, Zheng, J, Nourse, A, Kurinov, I, Peters, J.M, Stark, H, Schulman, B.A. | | Deposit date: | 2014-08-13 | | Release date: | 2014-10-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.755 Å) | | Cite: | Mechanism of Polyubiquitination by Human Anaphase-Promoting Complex: RING Repurposing for Ubiquitin Chain Assembly.
Mol.Cell, 56, 2014
|
|
2G3S
 
 | | RNA structure containing GU base pairs | | Descriptor: | 5'-R(*GP*GP*CP*GP*UP*GP*CP*C)-3', MAGNESIUM ION | | Authors: | Jang, S.B, Hung, L.W, Jeong, M.S, Holbrook, E.L, Chen, X, Turner, D.H, Holbrook, S.R. | | Deposit date: | 2006-02-20 | | Release date: | 2007-01-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | The crystal structure at 1.5 angstroms resolution of an RNA octamer duplex containing tandem G.U basepairs
Biophys.J., 90, 2006
|
|
5J3Y
 
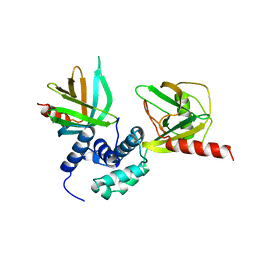 | | Crystal structure of S. pombe Dcp2:Dcp1 mRNA decapping complex | | Descriptor: | mRNA decapping complex subunit 2, mRNA-decapping enzyme subunit 1 | | Authors: | Valkov, E, Muthukumar, S, Chang, C.T, Jonas, S, Weichenrieder, O, Izaurralde, E. | | Deposit date: | 2016-03-31 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.288 Å) | | Cite: | Structure of the Dcp2-Dcp1 mRNA-decapping complex in the activated conformation.
Nat.Struct.Mol.Biol., 23, 2016
|
|
6S1Z
 
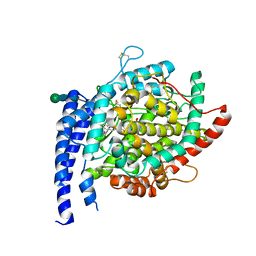 | | Crystal structure of Anopheles gambiae AnoACE2 in complex with fosinoprilat | | Descriptor: | Angiotensin-converting enzyme, ZINC ION, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cozier, G.E, Acharya, K.R, Cashman, J.S. | | Deposit date: | 2019-06-19 | | Release date: | 2019-11-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of angiotensin-converting enzyme from Anopheles gambiae in its native form and with a bound inhibitor.
Biochem.J., 476, 2019
|
|
8ERM
 
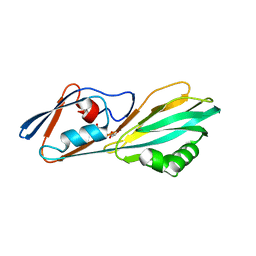 | | Crystal structure of FliC D2/D3 domains from Pseudomonas aeruginosa PAO1 | | Descriptor: | B-type flagellin, GLYCEROL, SULFATE ION | | Authors: | Nedeljkovic, M, Bonsor, D.A, Postel, S, Sundberg, E.J. | | Deposit date: | 2022-10-12 | | Release date: | 2023-05-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.475 Å) | | Cite: | An unbroken network of interactions connecting flagellin domains is required for motility in viscous environments.
Plos Pathog., 19, 2023
|
|
8F0G
 
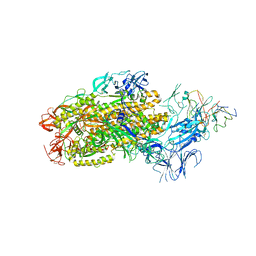 | | Structure of SARS-CoV-2 Omicron BA.1 spike in complex with antibody Fab 1C3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 1C3 Fab Heavy Chain, ... | | Authors: | Yu, X, Zyla, D, Hastie, K.M, Saphire, E.O. | | Deposit date: | 2022-11-02 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Potent Omicron-neutralizing antibodies isolated from a patient vaccinated 6 months before Omicron emergence.
Cell Rep, 42, 2023
|
|
8F0H
 
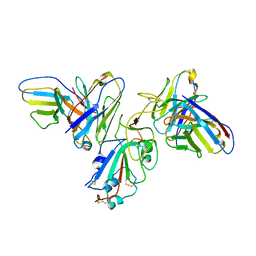 | | Structure of SARS-CoV-2 spike with antibody Fabs 2A10 and 1H2 (Local refinement of the RBD and Fabs 1H2 and 2A10) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Fab 1H2 heavy chain, Antibody Fab 1H2 light chain, ... | | Authors: | Yu, X, Zyla, D, Hastie, K.M, Saphire, E.O. | | Deposit date: | 2022-11-02 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Potent Omicron-neutralizing antibodies isolated from a patient vaccinated 6 months before Omicron emergence.
Cell Rep, 42, 2023
|
|
8EMM
 
 | |
8OVE
 
 | |
8OVA
 
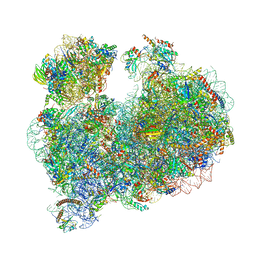 | |
7VFP
 
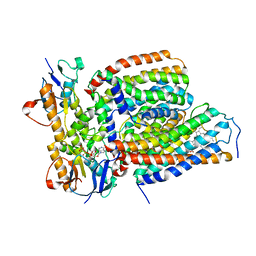 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli in complex with heme and single ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-09-13 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
6UYK
 
 | | Dark-operative protochlorophyllide oxidoreductase in the nucleotide-free form. | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase iron-sulfur ATP-binding protein | | Authors: | Bacik, J.P, Imran, S.M.S, Watkins, M.B, Corless, E, Antony, E, Ando, N. | | Deposit date: | 2019-11-13 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The flexible N-terminus of BchL autoinhibits activity through interaction with its [4Fe-4S] cluster and released upon ATP binding.
J.Biol.Chem., 296, 2020
|
|
