5RK1
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z1507502062 | | Descriptor: | 6-methoxy-1,3,4,5-tetrahydro-2H-1-benzazepin-2-one, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.271 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
2QMI
 
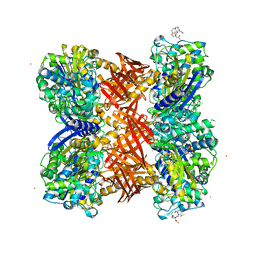 | | Structure of the octameric penicillin-binding protein homologue from Pyrococcus abyssi | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, LUTETIUM (III) ION, Pbp related beta-lactamase | | Authors: | Delfosse, V, Girard, E, Moulinier, L, Schultz, P, Mayer, C. | | Deposit date: | 2007-07-16 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the archaeal pab87 peptidase reveals a novel self-compartmentalizing protease family
Plos One, 4, 2009
|
|
5SPY
 
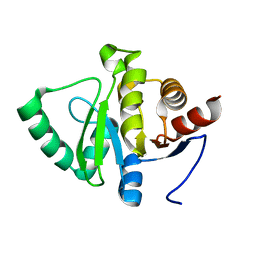 | |
5SQU
 
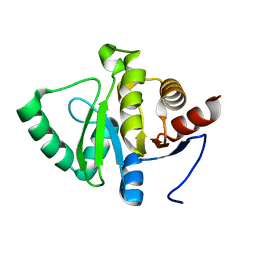 | |
6N95
 
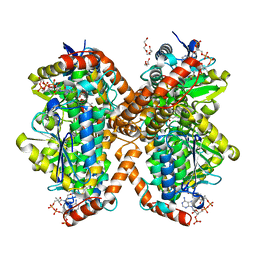 | | Methylmalonyl-CoA decarboxylase in complex with 2-sulfonate-propionyl-CoA | | Descriptor: | (2R)-sulfonatepropionyl-CoA, (2S)-sulfonatepropionyl-CoA, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Stunkard, L.M, Dixon, A.D, Huth, T.J, Lohman, J.R. | | Deposit date: | 2018-11-30 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Sulfonate/Nitro Bearing Methylmalonyl-Thioester Isosteres Applied to Methylmalonyl-CoA Decarboxylase Structure-Function Studies.
J. Am. Chem. Soc., 141, 2019
|
|
6MQ4
 
 | | GH5-4 broad specificity endoglucanase from Hungateiclostridium cellulolyticum | | Descriptor: | DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, ... | | Authors: | Bingman, C.A, Smith, R.W, Glasgow, E.M, Fox, B.G. | | Deposit date: | 2018-10-09 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A structural and kinetic survey of GH5_4 endoglucanases reveals determinants of broad substrate specificity and opportunities for biomass hydrolysis.
J.Biol.Chem., 295, 2020
|
|
5SRT
 
 | |
5SP9
 
 | |
5SP7
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010903509 - (S,S) isomer | | Descriptor: | (1S,2S)-1-[4-(cyclopropylcarbamamido)benzamido]-1,2,3,4-tetrahydronaphthalene-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1XSA
 
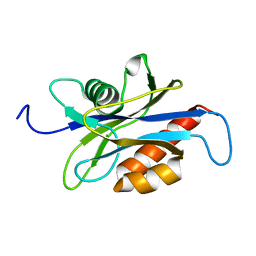 | | Structure of the nudix enzyme AP4A hydrolase from homo sapiens (E63A mutant) | | Descriptor: | Bis(5'-nucleosyl)-tetraphosphatase | | Authors: | Swarbrick, J.D, Buyya, S, Gunawardana, D, Gayler, K.R, McLennan, A.G, Gooley, P.R. | | Deposit date: | 2004-10-18 | | Release date: | 2004-12-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and Substrate-binding Mechanism of Human Ap4A Hydrolase
J.Biol.Chem., 280, 2005
|
|
5SPM
 
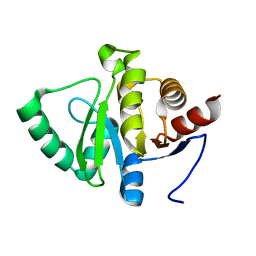 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with FRESH00002410346 | | Descriptor: | 4-hydroxy-6-(3-hydroxy-1-methyl-1,4,5,7-tetrahydro-6H-pyrazolo[3,4-c]pyridine-6-carbonyl)-2H-pyran-2-one, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
2DMR
 
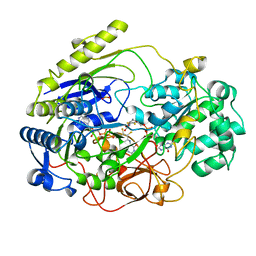 | | DITHIONITE REDUCED DMSO REDUCTASE FROM RHODOBACTER CAPSULATUS | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DMSO REDUCTASE, MOLYBDENUM(IV) ION, ... | | Authors: | Mcalpine, A.S, Bailey, S. | | Deposit date: | 1997-04-24 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molybdenum Active Centre of Dmso Reductase from Rhodobacter Capsulatus: Crystal Structure of the Oxidised Enzyme at 1.82-A Resolution and the Dithionite-Reduced Enzyme at 2.8-A Resolution
J.Biol.Inorg.Chem., 2, 1997
|
|
1RWT
 
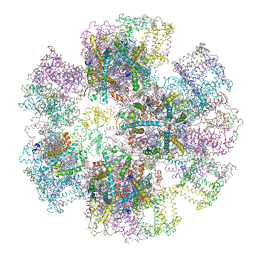 | | Crystal Structure of Spinach Major Light-harvesting complex at 2.72 Angstrom Resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Liu, Z, Yan, H, Wang, K, Kuang, T, Zhang, J, Gui, L, An, X, Chang, W. | | Deposit date: | 2003-12-17 | | Release date: | 2004-03-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal structure of spinach major light-harvesting complex at 2.72 A resolution
Nature, 428, 2004
|
|
2LUP
 
 | |
2LZK
 
 | | NMR solution structure of an N2-guanine DNA adduct derived from the potent tumorigen dibenzo[a,l]pyrene: Intercalation from the minor groove with ruptured Watson-Crick base pairing | | Descriptor: | (11S,12S,13S)-11,12,13,14-tetrahydronaphtho[1,2,3,4-pqr]tetraphene-11,12,13-triol, DNA (5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3') | | Authors: | Tang, Y, Liu, Z, Ding, S, Lin, C.H, Cai, Y, Rodriguez, F.A, Sayer, J.M, Jerina, D.M, Amin, S, Broyde, S, Geacintov, N.E. | | Deposit date: | 2012-10-04 | | Release date: | 2012-11-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Solution Structure of an N(2)-Guanine DNA Adduct Derived from the Potent Tumorigen Dibenzo[a,l]pyrene: Intercalation from the Minor Groove with Ruptured Watson-Crick Base Pairing.
Biochemistry, 51, 2012
|
|
5SJ4
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with 1-methyl-6-[2-(1-methyl-4-phenylimidazol-2-yl)ethyl]-5H-pyrazolo[3,4-d]pyrimidin-4-one | | Descriptor: | 1-methyl-6-{2-[(4R)-1-methyl-4-phenyl-4,5-dihydro-1H-imidazol-2-yl]ethyl}-1,2,3,5-tetrahydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Groebke-Zbinden, K, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
1WOP
 
 | | Crystal Structure of T-protein of the Glycine Cleavage System | | Descriptor: | Aminomethyltransferase, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid | | Authors: | Lee, H.H, Kim, D.J, Ahn, H.J, Ha, J.Y, Suh, S.W. | | Deposit date: | 2004-08-24 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of T-protein of the Glycine Cleavage System: Cofactor binding, insights into H-protein recognition, and molecular basis for understanding nonketotic hyperglycinemia
J.Biol.Chem., 279, 2004
|
|
2GTB
 
 | | Crystal structure of SARS coronavirus main peptidase (with an additional Ala at the N-terminus of each protomer) inhibited by an aza-peptide epoxide in the space group P43212 | | Descriptor: | (5S,8S,14R)-ETHYL 11-(3-AMINO-3-OXOPROPYL)-8-BENZYL-14-HYDROXY-5-ISOBUTYL-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,11-TETRAAZAPENTADECAN-15-OATE, 3C-like proteinase, ACETIC ACID | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C. | | Deposit date: | 2006-04-27 | | Release date: | 2006-12-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures Reveal an Induced-fit Binding of a Substrate-like Aza-peptide Epoxide to SARS Coronavirus Main Peptidase.
J.Mol.Biol., 366, 2007
|
|
2MIW
 
 | | Nuclear magnetic resonance studies of N2-guanine adducts derived from the tumorigen dibenzo[a,l]pyrene in DNA: Impact of adduct stereochemistry, size, and local DNA structure on solution conformations | | Descriptor: | (11R,12R,13R)-11,12,13,14-tetrahydronaphtho[1,2,3,4-pqr]tetraphene-11,12,13-triol, DNA_(5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3'), DNA_(5'-D(*GP*GP*TP*AP*GP*CP*GP*AP*TP*GP*G)-3') | | Authors: | Rodriguez, F.A, Liu, Z, Lin, C.H, Ding, S, Cai, Y, Kolbanovskiy, A, Kolbanovskiy, M, Amin, S, Broyde, S, Geacintov, N.E. | | Deposit date: | 2013-12-20 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Studies of an N(2)-Guanine Adduct Derived from the Tumorigen Dibenzo[a,l]pyrene in DNA: Impact of Adduct Stereochemistry, Size, and Local DNA Sequence on Solution Conformations.
Biochemistry, 53, 2014
|
|
2BEE
 
 | | Complex Between Paromomycin derivative JS4 and the 16S-Rrna A Site | | Descriptor: | (2S,3S,4R,5R,6R)-5-AMINO-2-(AMINOMETHYL)-6-((2R,3R,4R,5S)-4-(2-(3-AMINOPROPYLAMINO)ETHOXY)-5-((1R,2R,3S,5R,6S)-3,5-DIAM INO-2-((2S,3R,4R,5S,6R)-3-AMINO-4,5-DIHYDROXY-6-(HYDROXYMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-6-HYDROXYCYCLOHEXYLOXY)-2-( HYDROXYMETHYL)-TETRAHYDROFURAN-3-YLOXY)-TETRAHYDRO-2H-PYRAN-3,4-DIOL, 5'-R(*CP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3' | | Authors: | Francois, B, Westhof, E. | | Deposit date: | 2005-10-24 | | Release date: | 2005-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Antibacterial aminoglycosides with a modified mode of binding to the ribosomal-RNA decoding site
ANGEW.CHEM.INT.ED.ENGL., 43, 2004
|
|
2BYD
 
 | | Structure of aminoadipate-semialdehyde dehydrogenase- phosphopantetheinyl transferase | | Descriptor: | BROMIDE ION, HSPC223 | | Authors: | Bunkoczi, G, Wu, X, Dubinina, E, Johansson, C, Smee, C, Turnbull, A, Oppermann, U, von Delft, F, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J. | | Deposit date: | 2005-07-29 | | Release date: | 2005-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism and substrate recognition of human holo ACP synthase.
Chem. Biol., 14, 2007
|
|
2BE0
 
 | | Complex Between Paromomycin Derivative JS5-39 and the 16S-Rrna A-Site. | | Descriptor: | (2S,3S,4R,5R,6R)-5-AMINO-2-(AMINOMETHYL)-6-((2R,3R,4R,5S)-5-((1R,2R,3S,5R,6S)-3,5-DIAMINO-2-((2S,3R,4R,5S,6R)-3-AMINO-4 ,5-DIHYDROXY-6-(HYDROXYMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-6-HYDROXYCYCLOHEXYLOXY)-2-(HYDROXYMETHYL)-4-(2-((R)-PIPERIDI N-3-YLMETHYLAMINO)ETHOXY)-TETRAHYDROFURAN-3-YLOXY)-TETRAHYDRO-2H-PYRAN-3,4-DIOL, 5'-R(*CP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3' | | Authors: | Francois, B, Westhof, E. | | Deposit date: | 2005-10-21 | | Release date: | 2005-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Antibacterial aminoglycosides with a modified mode of binding to the ribosomal-RNA decoding site
ANGEW.CHEM.INT.ED.ENGL., 43, 2004
|
|
2MIV
 
 | | NMR studies of N2-guanine adducts derived from the tumorigen dibenzo[a,l]pyrene in DNA: Impact of adduct stereochemistry, size, and local DNA structure on solution conformations | | Descriptor: | (11R,12R,13R)-11,12,13,14-tetrahydronaphtho[1,2,3,4-pqr]tetraphene-11,12,13-triol, DNA_(5'-D(*CP*CP*AP*TP*CP*GP*CP*TP*AP*CP*C)-3'), DNA_(5'-D(*GP*GP*TP*AP*GP*GP*AP*TP*GP*G)-3') | | Authors: | Rodriguez, F.A, Liu, Z, Lin, C.H, Ding, S, Cai, Y, Kolbanovskiy, A, Kolbanovskiy, M, Amin, S, Broyde, S, Geacintov, N.E. | | Deposit date: | 2013-12-20 | | Release date: | 2014-04-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Studies of an N(2)-Guanine Adduct Derived from the Tumorigen Dibenzo[a,l]pyrene in DNA: Impact of Adduct Stereochemistry, Size, and Local DNA Sequence on Solution Conformations.
Biochemistry, 53, 2014
|
|
4LZF
 
 | | A novel domain in the microcephaly protein CPAP suggests a role in centriole architecture | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Centrosomal P4.1-associated protein, SCL-interrupting locus protein homolog, ... | | Authors: | Hatzopoulos, G.N, Erat, M.C, Cutts, E, Rogala, K, Slatter, L, Stansfeld, P.J, Vakonakis, I. | | Deposit date: | 2013-07-31 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structural analysis of the G-box domain of the microcephaly protein CPAP suggests a role in centriole architecture.
Structure, 21, 2013
|
|
3EJK
 
 | |
