1HQ0
 
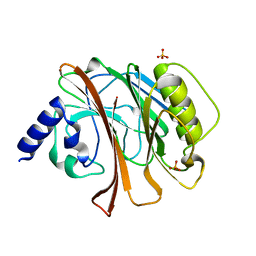 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF E.COLI CYTOTOXIC NECROTIZING FACTOR TYPE 1 | | Descriptor: | CYTOTOXIC NECROTIZING FACTOR 1, PHOSPHATE ION | | Authors: | Buetow, L, Flatau, G, Chiu, K, Boquet, P, Ghosh, P. | | Deposit date: | 2000-12-13 | | Release date: | 2001-07-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of the Rho-activating domain of Escherichia coli cytotoxic necrotizing factor 1.
Nat.Struct.Biol., 8, 2001
|
|
3K7T
 
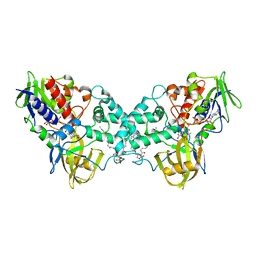 | | Crystal structure of apo-form 6-hydroxy-L-nicotine oxidase, crystal form P3121 | | Descriptor: | (1R)-2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-1-[(pentadecanoyloxy)methyl]ethyl (12E)-hexadeca-9,12-dienoate, 6-hydroxy-L-nicotine oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Bourenkov, G.P, Kachalova, G.S, Bartunik, H.D. | | Deposit date: | 2009-10-13 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure Analysis of Free and Substrate-Bound 6-Hydroxy-l-Nicotine Oxidase from Arthrobacter nicotinovorans.
J.Mol.Biol., 396, 2010
|
|
1HZG
 
 | | CRYSTAL STRUCTURE OF THE INACTIVE C866S MUTANT OF THE CATALYTIC DOMAIN OF E. COLI CYTOTOXIC NECROTIZING FACTOR 1 | | Descriptor: | CYTOTOXIC NECROTIZING FACTOR 1, PHOSPHATE ION | | Authors: | Buetow, L, Flatau, G, Chiu, K, Boquet, P, Ghosh, P. | | Deposit date: | 2001-01-24 | | Release date: | 2001-07-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of the Rho-activating domain of Escherichia coli cytotoxic necrotizing factor 1.
Nat.Struct.Biol., 8, 2001
|
|
1OVA
 
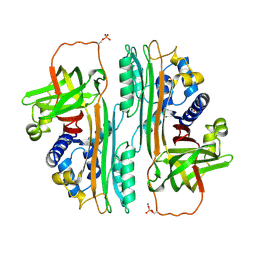 | |
4RNU
 
 | | G303 Circular Permutation of Old Yellow Enzyme | | Descriptor: | FLAVIN MONONUCLEOTIDE, NADPH dehydrogenase 1, PHOSPHATE ION | | Authors: | Horton, J.R, Daugherty, A.B, Cheng, X, Lutz, S. | | Deposit date: | 2014-10-26 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.677 Å) | | Cite: | STRUCTURAL AND FUNCTIONAL CONSEQUENCES OF CIRCULAR PERMUTATION ON THE ACTIVE SITE OF OLD YELLOW ENZYME.
ACS Catal, 5, 2015
|
|
6DHZ
 
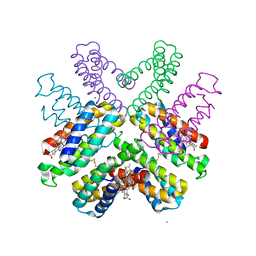 | |
1GW2
 
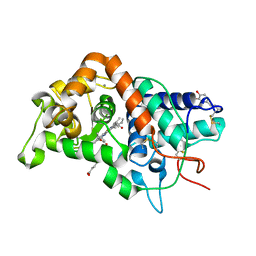 | | RECOMBINANT HORSERADISH PEROXIDASE C1A THR171SER IN COMPLEX WITH FERULIC ACID | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, CALCIUM ION, PEROXIDASE C1A, ... | | Authors: | Henriksen, A, Meno, K, Brissett, N, Gajhede, M. | | Deposit date: | 2002-03-03 | | Release date: | 2003-03-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Hrpc Heme Crevice Architecture
To be Published
|
|
5E4X
 
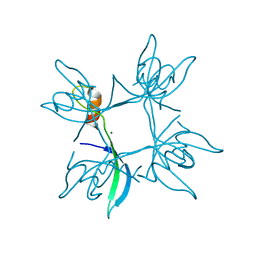 | | Crystal structure of cpSRP43 chromodomain 3 | | Descriptor: | MAGNESIUM ION, Signal recognition particle 43 kDa protein, chloroplastic | | Authors: | Horn, A, Ahmed, Y.L, Wild, K, Sinning, I. | | Deposit date: | 2015-10-07 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for cpSRP43 chromodomain selectivity and dynamics in Alb3 insertase interaction.
Nat Commun, 6, 2015
|
|
1KAQ
 
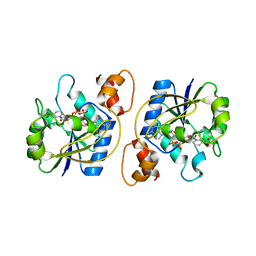 | | Structure of Bacillus subtilis Nicotinic Acid Mononucleotide Adenylyl Transferase | | Descriptor: | NICOTINATE-NUCLEOTIDE ADENYLYLTRANSFERASE, NICOTINIC ACID ADENINE DINUCLEOTIDE | | Authors: | Olland, A.M, Underwood, K.W, Czerwinski, R.M, Lo, M.C, Aulabaugh, A, Bard, J, Stahl, M.L, Somers, W.S, Sullivan, F.X, Chopra, R. | | Deposit date: | 2001-11-02 | | Release date: | 2002-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Identification, characterization, and crystal structure of Bacillus subtilis nicotinic acid mononucleotide adenylyltransferase.
J.Biol.Chem., 277, 2002
|
|
7S2S
 
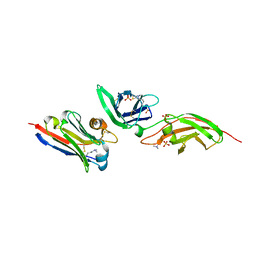 | | nanobody bound to Interleukin-2Rbeta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, IL2Rb-binding nanobody, Interleukin-2 receptor subunit beta, ... | | Authors: | Glassman, C.R, Jude, K.M, Yen, M, Garcia, K.C. | | Deposit date: | 2021-09-03 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Facile discovery of surrogate cytokine agonists.
Cell, 185, 2022
|
|
7S2R
 
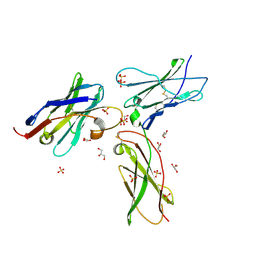 | | nanobody bound to IL-2Rg | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokine receptor common subunit gamma, ... | | Authors: | Glassman, C.R, Jude, K.M, Yen, M, Garcia, K.C. | | Deposit date: | 2021-09-03 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Facile discovery of surrogate cytokine agonists.
Cell, 185, 2022
|
|
1O87
 
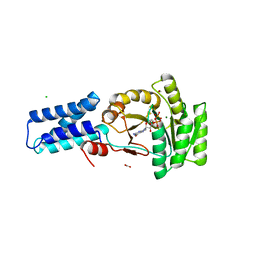 | | A new MgGDP complex of the Ffh NG domain | | Descriptor: | CHLORIDE ION, FORMIC ACID, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Freymann, D.M. | | Deposit date: | 2002-11-25 | | Release date: | 2004-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel Protein and Mg2+ Configurations in the Mg2+Gdp Complex of the Srp Gtpase Ffh
Proteins: Struct.,Funct., Genet., 54, 2004
|
|
8DGK
 
 | |
5AYY
 
 | | CRYSTAL STRUCTURE OF HUMAN QUINOLINATE PHOSPHORIBOSYLTRANSFERASE IN COMPLEX WITH THE REACTANT QUINOLINATE | | Descriptor: | Nicotinate-nucleotide pyrophosphorylase [carboxylating], QUINOLINIC ACID | | Authors: | Youn, H.S, Kim, T.G, Kim, M.K, Kang, G.B, Kang, J.Y, Seo, Y.J, Lee, J.G, An, J.Y, Park, K.R, Lee, Y, Im, Y.J, Lee, J.H, Fukuoka, S.I, Eom, S.H. | | Deposit date: | 2015-09-14 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structural Insights into the Quaternary Catalytic Mechanism of Hexameric Human Quinolinate Phosphoribosyltransferase, a Key Enzyme in de novo NAD Biosynthesis
Sci Rep, 6, 2016
|
|
1GQV
 
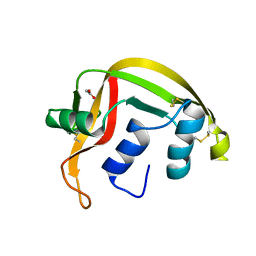 | | Atomic Resolution (0.98A) Structure of Eosinophil-Derived Neurotoxin | | Descriptor: | ACETATE ION, EOSINOPHIL-DERIVED NEUROTOXIN | | Authors: | Swaminathan, G.J, Holloway, D.E, Veluraja, K, Acharya, K.R. | | Deposit date: | 2001-12-05 | | Release date: | 2002-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Atomic Resolution (0.98 A) Structure of Eosinophil-Derived Neurotoxin
Biochemistry, 41, 2002
|
|
5B26
 
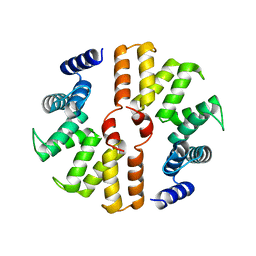 | | Crystal structure of mouse SEL1L | | Descriptor: | Protein sel-1 homolog 1 | | Authors: | Jeong, H, Lee, C. | | Deposit date: | 2016-01-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of SEL1L: Insight into the roles of SLR motifs in ERAD pathway
Sci Rep, 6, 2016
|
|
3K8J
 
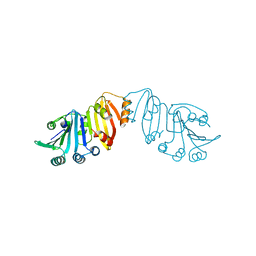 | | Structure of crystal form III of TP0453 | | Descriptor: | 30kLP | | Authors: | Zhu, G, Luthra, A, Desrosiers, D, Koszelak-Rosenblum, M, Mulay, V, Radolf, J.D, Malkowski, M.G. | | Deposit date: | 2009-10-14 | | Release date: | 2010-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Transition from Closed to Open Conformation of Treponema pallidum Outer Membrane-associated Lipoprotein TP0453 Involves Membrane Sensing and Integration by Two Amphipathic Helices.
J.Biol.Chem., 286, 2011
|
|
1O7B
 
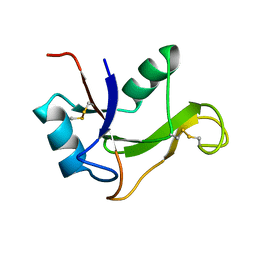 | | Refined solution structure of the human TSG-6 Link module | | Descriptor: | TUMOR NECROSIS FACTOR-INDUCIBLE PROTEIN TSG-6 | | Authors: | Blundell, C.D, Teriete, P, Kahmann, J.D, Pickford, A.R, Campbell, I.D, Day, A.J. | | Deposit date: | 2002-10-29 | | Release date: | 2003-10-23 | | Last modified: | 2018-01-24 | | Method: | SOLUTION NMR | | Cite: | The link module from ovulation- and inflammation-associated protein TSG-6 changes conformation on hyaluronan binding.
J. Biol. Chem., 278, 2003
|
|
1RKU
 
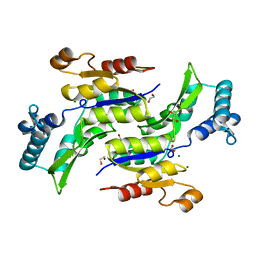 | | Crystal Structure of ThrH gene product of Pseudomonas Aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, homoserine kinase | | Authors: | Singh, S.K, Yang, K, Subramanian, K, Karthikeyan, S, Huynh, T, Zhang, X, Phillips, M.A, Zhang, H. | | Deposit date: | 2003-11-23 | | Release date: | 2004-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The thrH Gene Product of Pseudomonas aeruginosa Is a Dual Activity Enzyme with a Novel Phosphoserine:Homoserine Phosphotransferase Activity.
J.Biol.Chem., 279, 2004
|
|
6DHY
 
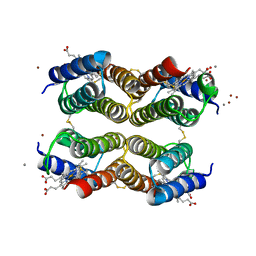 | |
1KRH
 
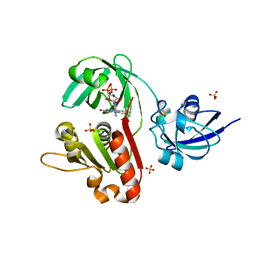 | | X-ray Structure of Benzoate Dioxygenase Reductase | | Descriptor: | Benzoate 1,2-Dioxygenase Reductase, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Karlsson, A, Beharry, Z.M, Eby, D.M, Coulter, E.D, Niedle, E.L, Kurtz Jr, D.M, Eklund, H, Ramaswamy, S. | | Deposit date: | 2002-01-09 | | Release date: | 2002-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystal structure of benzoate 1,2-dioxygenase reductase from Acinetobacter sp. strain ADP1.
J.Mol.Biol., 318, 2002
|
|
5CX1
 
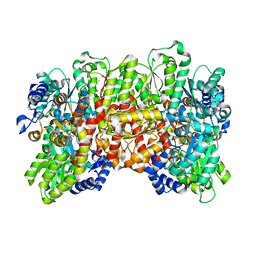 | | Nitrogenase molybdenum-iron protein beta-K400E mutant | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Owens, C.P, Luca, M.A, Tezcan, F.A. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7476 Å) | | Cite: | Evidence for Functionally Relevant Encounter Complexes in Nitrogenase Catalysis.
J.Am.Chem.Soc., 137, 2015
|
|
1KET
 
 | | The crystal structure of dTDP-D-glucose 4,6-dehydratase (RmlB) from Streptococcus suis with thymidine diphosphate bound | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, THYMIDINE-5'-DIPHOSPHATE, dTDP-D-glucose 4,6-dehydratase | | Authors: | Allard, S.T.M, Beis, K, Giraud, M.-F, Hegeman, A.D, Gross, J.W, Whitfield, C, Graninger, M, Messner, P, Allen, A.G, Naismith, J.H. | | Deposit date: | 2001-11-17 | | Release date: | 2002-01-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward a structural understanding of the dehydratase mechanism.
Structure, 10, 2002
|
|
5D70
 
 | | Crystal structure of MOR03929, a neutralizing anti-human GM-CSF antibody Fab fragment in complex with human GM-CSF | | Descriptor: | Granulocyte-macrophage colony-stimulating factor, Immunglobulin G1 Fab fragment, heavy chain, ... | | Authors: | Eylenstein, R, Weinfurtner, D, Steidl, S, Boettcher, J, Augustin, M. | | Deposit date: | 2015-08-13 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Molecular basis of in vitro affinity maturation and functional evolution of a neutralizing anti-human GM-CSF antibody.
Mabs, 8, 2016
|
|
5D7S
 
 | | Crystal structure of MOR04357, a neutralizing anti-human GM-CSF antibody Fab fragment | | Descriptor: | (2S,3S)-butane-2,3-diol, Immunglobulin G1 Fab fragment, heavy chain, ... | | Authors: | Eylenstein, R, Weinfurtner, D, Steidl, S, Boettcher, J, Augustin, M. | | Deposit date: | 2015-08-14 | | Release date: | 2015-10-14 | | Last modified: | 2016-01-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Molecular basis of in vitro affinity maturation and functional evolution of a neutralizing anti-human GM-CSF antibody.
Mabs, 8, 2016
|
|
