6OO7
 
 | | Cryo-EM structure of the C2-symmetric TRPV2/RTx complex in nanodiscs | | Descriptor: | TRPV2, resiniferatoxin | | Authors: | Zubcevic, L, Hsu, A.L, Borgnia, M.J, Lee, S.-Y. | | Deposit date: | 2019-04-22 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Symmetry transitions during gating of the TRPV2 ion channel in lipid membranes.
Elife, 8, 2019
|
|
1KVZ
 
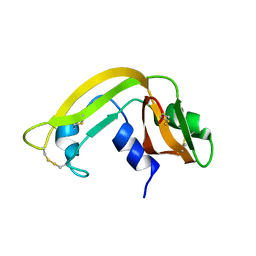 | | Solution Structure of Cytotoxic RC-RNase4 | | Descriptor: | RC-RNase4 | | Authors: | Hsu, C.-H, Liao, Y.-D, Chen, L.-W, Wu, S.-H, Chen, C. | | Deposit date: | 2002-01-28 | | Release date: | 2002-07-28 | | Last modified: | 2022-12-21 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Cytotoxic RNase 4 from the Oocytes of Bullfrog Rana Catesbeiana
J.MOL.BIOL., 326, 2003
|
|
1KEU
 
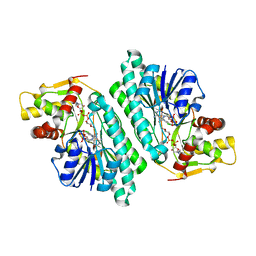 | | The crystal structure of dTDP-D-glucose 4,6-dehydratase (RmlB) from Salmonella enterica serovar Typhimurium with dTDP-D-glucose bound | | Descriptor: | 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, dTDP-D-glucose 4,6-dehydratase | | Authors: | Allard, S.T.M, Beis, K, Giraud, M.-F, Hegeman, A.D, Gross, J.W, Whitfield, C, Graninger, M, Messner, P, Allen, A.G, Naismith, J.H. | | Deposit date: | 2001-11-17 | | Release date: | 2002-01-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Toward a structural understanding of the dehydratase mechanism.
Structure, 10, 2002
|
|
6OO5
 
 | | Cryo-EM structure of the C2-symmetric TRPV2/RTx complex in amphipol resolved to 4.2 A | | Descriptor: | TRPV2, resiniferatoxin | | Authors: | Zubcevic, L, Hsu, A.L, Borgnia, M.J, Lee, S.-Y. | | Deposit date: | 2019-04-22 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Symmetry transitions during gating of the TRPV2 ion channel in lipid membranes.
Elife, 8, 2019
|
|
1KEP
 
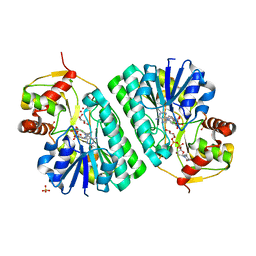 | | The crystal structure of dTDP-D-glucose 4,6-dehydratase (RmlB) from Streptococcus suis with dTDP-xylose bound | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, THYMIDINE-5'-DIPHOSPHO-BETA-D-XYLOSE, ... | | Authors: | Allard, S.T.M, Beis, K, Giraud, M.-F, Hegeman, A.D, Gross, J.W, Whitfield, C, Graninger, M, Messner, P, Allen, A.G, Naismith, J.H. | | Deposit date: | 2001-11-16 | | Release date: | 2002-01-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward a structural understanding of the dehydratase mechanism.
Structure, 10, 2002
|
|
6OP1
 
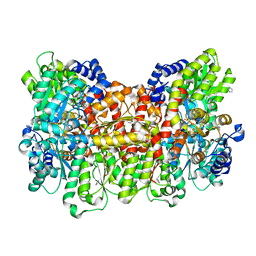 | |
1KS9
 
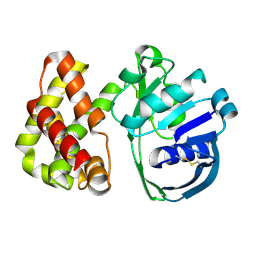 | | Ketopantoate Reductase from Escherichia coli | | Descriptor: | 2-DEHYDROPANTOATE 2-REDUCTASE | | Authors: | Matak-Vinkovic, D, Vinkovic, M, Saldanha, S.A, Ashurst, J.A, von Delft, F, Inoue, T, Miguel, R.N, Smith, A.G, Blundell, T.L, Abell, C. | | Deposit date: | 2002-01-11 | | Release date: | 2002-01-25 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Escherichia coli ketopantoate reductase at 1.7 A resolution and insight into the enzyme mechanism.
Biochemistry, 40, 2001
|
|
1JCR
 
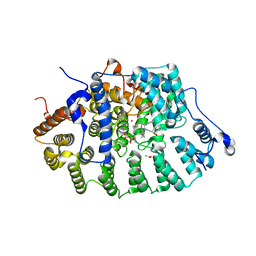 | | CRYSTAL STRUCTURE OF RAT PROTEIN FARNESYLTRANSFERASE COMPLEXED WITH THE NON-SUBSTRATE TETRAPEPTIDE INHIBITOR CVFM AND FARNESYL DIPHOSPHATE SUBSTRATE | | Descriptor: | ACETIC ACID, FARNESYL DIPHOSPHATE, PROTEIN FARNESYLTRANSFERASE, ... | | Authors: | Long, S.B, Casey, P.J, Beese, L.S. | | Deposit date: | 2001-06-11 | | Release date: | 2001-11-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of human protein farnesyltransferase reveals the basis for inhibition by CaaX tetrapeptides and their mimetics.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
5XXR
 
 | |
3KJO
 
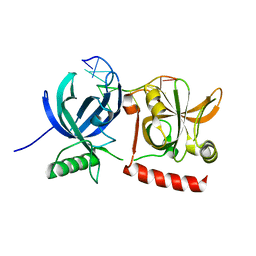 | | Crystal Structure of hPOT1V2-dTrUd(AGGGTTAG) | | Descriptor: | DNA/RNA (5'-D(*T)-R(P*U)-D(P*AP*GP*GP*GP*TP*TP*AP*G)-3'), Protection of telomeres protein 1 | | Authors: | Nandakumar, J, Cech, T.R, Podell, E.R. | | Deposit date: | 2009-11-03 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | How telomeric protein POT1 avoids RNA to achieve specificity for single-stranded DNA.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1JCQ
 
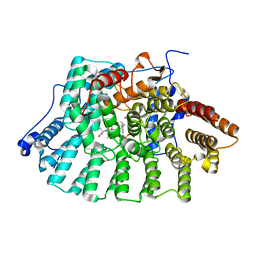 | | CRYSTAL STRUCTURE OF HUMAN PROTEIN FARNESYLTRANSFERASE COMPLEXED WITH FARNESYL DIPHOSPHATE AND THE PEPTIDOMIMETIC INHIBITOR L-739,750 | | Descriptor: | 2(S)-{2(S)-[2(R)-AMINO-3-MERCAPTO]PROPYLAMINO-3(S)-METHYL}PENTYLOXY-3-PHENYLPROPIONYLMETHIONINE SULFONE, ACETIC ACID, FARNESYL DIPHOSPHATE, ... | | Authors: | Long, S.B, Casey, P.J, Beese, L.S. | | Deposit date: | 2001-06-11 | | Release date: | 2001-11-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of human protein farnesyltransferase reveals the basis for inhibition by CaaX tetrapeptides and their mimetics.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1GWU
 
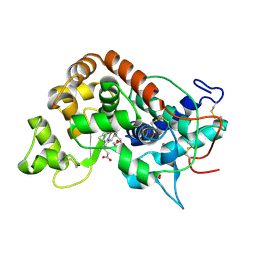 | |
3KF3
 
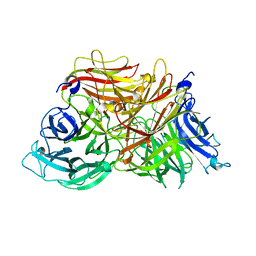 | |
1YQO
 
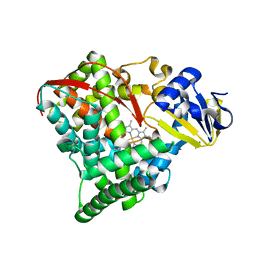 | | T268A mutant heme domain of flavocytochrome P450 BM3 | | Descriptor: | Bifunctional P-450:NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Clark, J.P, Miles, C.S, Mowat, C.G, Walkinshaw, M.D, Reid, G.A, Daff, S.N, Chapman, S.K. | | Deposit date: | 2005-02-02 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of Thr268 and Phe393 in cytochrome P450 BM3.
J.Inorg.Biochem., 100, 2006
|
|
5D71
 
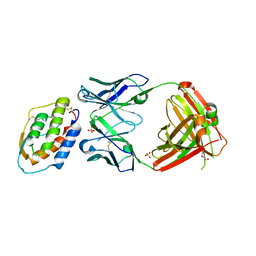 | | Crystal structure of MOR04302, a neutralizing anti-human GM-CSF antibody Fab fragment in complex with human GM-CSF | | Descriptor: | DI(HYDROXYETHYL)ETHER, Granulocyte-macrophage colony-stimulating factor, Immunglobulin G1 Fab fragment, ... | | Authors: | Eylenstein, R, Weinfurtner, D, Steidl, S, Boettcher, J, Augustin, M. | | Deposit date: | 2015-08-13 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Molecular basis of in vitro affinity maturation and functional evolution of a neutralizing anti-human GM-CSF antibody.
Mabs, 8, 2016
|
|
1O7C
 
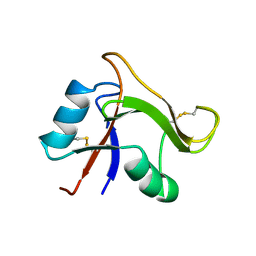 | | Solution structure of the human TSG-6 Link module in the presence of a hyaluronan octasaccharide | | Descriptor: | TUMOR NECROSIS FACTOR-INDUCIBLE PROTEIN TSG-6 | | Authors: | Blundell, C.D, Teriete, P, Kahmann, J.D, Pickford, A.R, Campbell, I.D, Day, A.J. | | Deposit date: | 2002-10-29 | | Release date: | 2003-10-23 | | Last modified: | 2018-01-24 | | Method: | SOLUTION NMR | | Cite: | The link module from ovulation- and inflammation-associated protein TSG-6 changes conformation on hyaluronan binding.
J. Biol. Chem., 278, 2003
|
|
6OP4
 
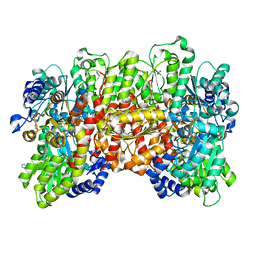 | | Selenium-incorporated, carbon monoxide-inhibited, reactivated FeMo-cofactor of nitrogenase from Azotobacter vinelandii | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Arias, R.J, Rees, D.C. | | Deposit date: | 2019-04-24 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Localized Electronic Structure of Nitrogenase FeMoco Revealed by Selenium K-Edge High Resolution X-ray Absorption Spectroscopy.
J.Am.Chem.Soc., 141, 2019
|
|
5CSS
 
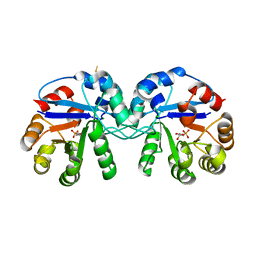 | | Crystal structure of triosephosphate isomerase from Thermoplasma acidophilum with glycerol 3-phosphate | | Descriptor: | CHLORIDE ION, SN-GLYCEROL-3-PHOSPHATE, Triosephosphate isomerase | | Authors: | Park, S.H, Kim, H.S, Song, M.K, Kim, K.R, Park, J.S, Han, B.W. | | Deposit date: | 2015-07-23 | | Release date: | 2016-06-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure and Stability of the Dimeric Triosephosphate Isomerase from the Thermophilic Archaeon Thermoplasma acidophilum.
Plos One, 10, 2015
|
|
5D72
 
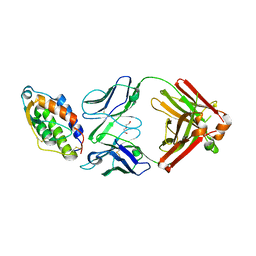 | | Crystal structure of MOR04252, a neutralizing anti-human GM-CSF antibody Fab fragment in complex with human GM-CSF | | Descriptor: | DI(HYDROXYETHYL)ETHER, Granulocyte-macrophage colony-stimulating factor, Immunglobulin G1 Fab fragment, ... | | Authors: | Eylenstein, R, Weinfurtner, D, Steidl, S, Boettcher, J, Augustin, M. | | Deposit date: | 2015-08-13 | | Release date: | 2015-10-14 | | Last modified: | 2016-01-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis of in vitro affinity maturation and functional evolution of a neutralizing anti-human GM-CSF antibody.
Mabs, 8, 2016
|
|
2A6P
 
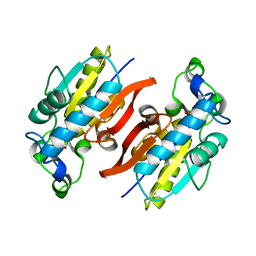 | | Structure Solution to 2.2 Angstrom and Functional Characterisation of the Open Reading Frame Rv3214 from Mycobacterium tuberculosis | | Descriptor: | GLYCEROL, POSSIBLE PHOSPHOGLYCERATE MUTASE GPM2, SULFATE ION | | Authors: | Watkins, H.A, Yu, M, Baker, E.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-07-03 | | Release date: | 2006-05-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Analysis of Rv3214 from Mycobacterium tuberculosis, a Protein with Conflicting Functional Annotations, Leads to Its Characterization as a Phosphatase.
J.Bacteriol., 188, 2006
|
|
1M55
 
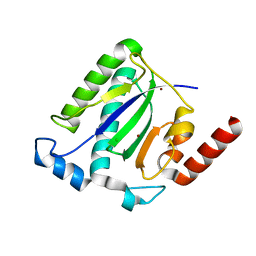 | | Catalytic domain of the Adeno Associated Virus type 5 Rep protein | | Descriptor: | CHLORIDE ION, Rep protein, ZINC ION | | Authors: | Hickman, A.B, Ronning, D.R, Kotin, R.M, Dyda, F. | | Deposit date: | 2002-07-08 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural unity among viral origin binding proteins: crystal structure of the nuclease domain of adeno-associated virus Rep.
Mol.Cell, 10, 2002
|
|
1HKY
 
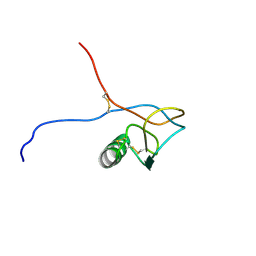 | | Solution structure of a PAN module from Eimeria tenella | | Descriptor: | MICRONEME PROTEIN 5 PRECURSOR | | Authors: | Brown, P.J, Mulvey, D, Potts, J.R, Tomley, F.M, Campbell, I.D. | | Deposit date: | 2002-10-03 | | Release date: | 2002-10-17 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a Pan Module from the Apicomplexan Parasite Eimeria Tenella
J.Struct.Funct.Genom., 4, 2003
|
|
5D1P
 
 | | Archaeal ATP-dependent RNA ligase - form 2 | | Descriptor: | ATP-dependent RNA ligase, MAGNESIUM ION, SULFATE ION | | Authors: | Murakami, K.S. | | Deposit date: | 2015-08-04 | | Release date: | 2016-03-09 | | Last modified: | 2016-03-30 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Structural and mutational analysis of archaeal ATP-dependent RNA ligase identifies amino acids required for RNA binding and catalysis.
Nucleic Acids Res., 44, 2016
|
|
1M44
 
 | | Aminoglycoside 2'-N-acetyltransferase from Mycobacterium tuberculosis-APO Structure | | Descriptor: | Aminoglycoside 2'-N-acetyltransferase, SULFATE ION | | Authors: | Vetting, M.W, Hegde, S.S, Javid-Majd, F, Blanchard, J.S, Roderick, S.L. | | Deposit date: | 2002-07-02 | | Release date: | 2002-08-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Aminoglycoside 2'-N-acetyltransferase from Mycobacterium tuberculosis in complex with coenzyme A and aminoglycoside substrates.
Nat.Struct.Biol., 9, 2002
|
|
3K8H
 
 | | Structure of crystal form I of TP0453 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 30kLP | | Authors: | Zhu, G, Luthra, A, Desrosiers, D, Koszelak-Rosenblum, M, Mulay, V, Radolf, J.D, Malkowski, M.G. | | Deposit date: | 2009-10-14 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | The Transition from Closed to Open Conformation of Treponema pallidum Outer Membrane-associated Lipoprotein TP0453 Involves Membrane Sensing and Integration by Two Amphipathic Helices.
J.Biol.Chem., 286, 2011
|
|
