5DFI
 
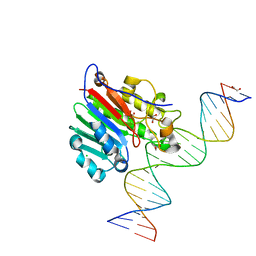 | | Human APE1 phosphorothioate substrate complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*(OMC)P*(48Z)P*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | Authors: | Freudenthal, B.D, Wilson, S.H. | | Deposit date: | 2015-08-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Capturing snapshots of APE1 processing DNA damage.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5DFH
 
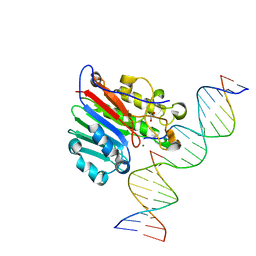 | | Human APE1 mismatch product complex | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*T)-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Freudenthal, B.D, Wilson, S.H. | | Deposit date: | 2015-08-26 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Capturing snapshots of APE1 processing DNA damage.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5CW1
 
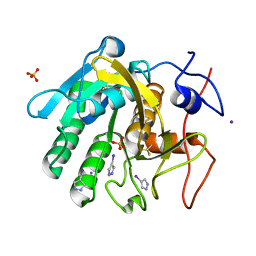 | | Proteinase K complexed with 4-iodopyrazole | | Descriptor: | 4-IODOPYRAZOLE, IODIDE ION, Proteinase K, ... | | Authors: | Bauman, J.D, Arnold, E. | | Deposit date: | 2015-07-27 | | Release date: | 2015-12-30 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Rapid experimental SAD phasing and hot spot identification with halogenated fragments
Iucrj, 3, 2016
|
|
5DCG
 
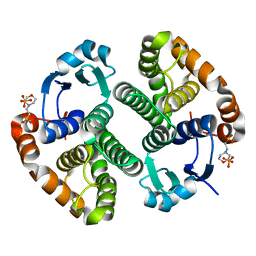 | |
8T5L
 
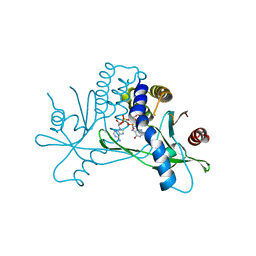 | | Crystal structure of STING CTD in complex with 2'3'-cGAMP | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Li, Y, Li, P, Sun, D. | | Deposit date: | 2023-06-13 | | Release date: | 2023-07-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and Biological Evaluations of a Non-Nucleoside STING Agonist Specific for Human STING A230 Variants.
Biorxiv, 2023
|
|
5DFP
 
 | | Crystal structure of PAK1 in complex with an inhibitor compound FRAX1036 | | Descriptor: | 6-[2-chloro-4-(6-methylpyrazin-2-yl)phenyl]-8-ethyl-2-{[2-(1-methylpiperidin-4-yl)ethyl]amino}pyrido[2,3-d]pyrimidin-7(8H)-one, DIMETHYL SULFOXIDE, Serine/threonine-protein kinase PAK 1 | | Authors: | Maksimoska, J, Marmorstein, R, Wang, W. | | Deposit date: | 2015-08-27 | | Release date: | 2016-01-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design of Selective PAK1 Inhibitor G-5555: Improving Properties by Employing an Unorthodox Low-pK a Polar Moiety.
Acs Med.Chem.Lett., 6, 2015
|
|
2CCK
 
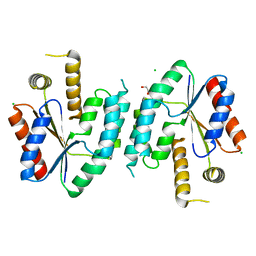 | | CRYSTAL STRUCTURE OF UNLIGANDED S. AUREUS THYMIDYLATE KINASE | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, THYMIDYLATE KINASE | | Authors: | Kotaka, M, Dhaliwal, B, Ren, J, Nichols, C.E, Angell, R, Lockyer, M, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2006-01-16 | | Release date: | 2006-03-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structures of S. Aureus Thymidylate Kinase Reveal an Atypical Active Site Configuration and an Intermediate Conformational State Upon Substrate Binding
Protein Sci., 15, 2006
|
|
5DG0
 
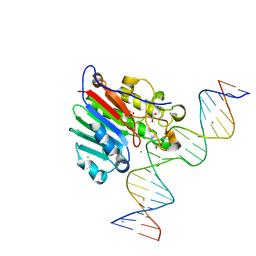 | |
5DJL
 
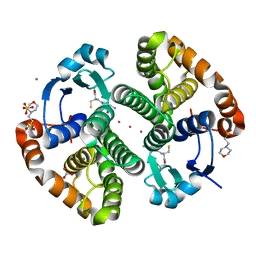 | |
5DAI
 
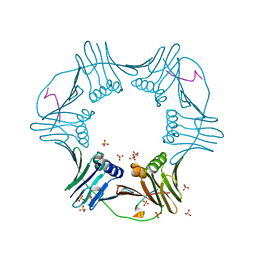 | |
2PTA
 
 | | PANDINUS TOXIN K-A (PITX-KA) FROM PANDINUS IMPERATOR, NMR, 20 STRUCTURES | | Descriptor: | PANDINUS TOXIN K-ALPHA | | Authors: | Tenenholz, T.C, Rogowski, R.S, Collins, J.H, Blaustein, M.P, Weber, D.J. | | Deposit date: | 1996-11-26 | | Release date: | 1997-12-10 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure for Pandinus toxin K-alpha (PiTX-K alpha), a selective blocker of A-type potassium channels.
Biochemistry, 36, 1997
|
|
8SLV
 
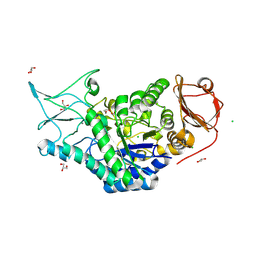 | | Structure of a salivary alpha-glucosidase from the mosquito vector Aedes aegypti. | | Descriptor: | 1,3-PROPANDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gittis, A.G, Williams, A.E, Garboczi, D, Calvo, E. | | Deposit date: | 2023-04-24 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural and functional comparisons of salivary alpha-glucosidases from the mosquito vectors Aedes aegypti, Anopheles gambiae, and Culex quinquefasciatus.
Insect Biochem.Mol.Biol., 167, 2024
|
|
5DFF
 
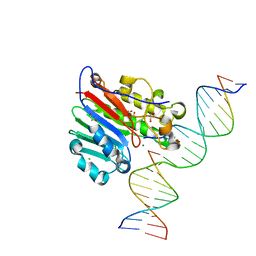 | | Human APE1 product complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Freudenthal, B.D, Wilson, S.H. | | Deposit date: | 2015-08-26 | | Release date: | 2015-10-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Capturing snapshots of APE1 processing DNA damage.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5D1K
 
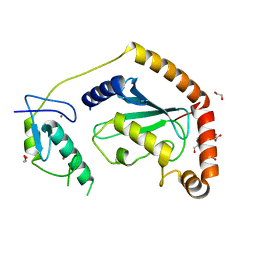 | | Crystal Structure of UbcH5B in Complex with the RING-U5BR Fragment of AO7 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase RNF25, ... | | Authors: | Liang, Y.-H, Li, S, Weissman, A.M, Ji, X. | | Deposit date: | 2015-08-04 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Insights into Ubiquitination from the Unique Clamp-like Binding of the RING E3 AO7 to the E2 UbcH5B.
J.Biol.Chem., 290, 2015
|
|
8STY
 
 | |
8SSY
 
 | |
5DD0
 
 | | Crystal structures in an anti-HIV antibody lineage from immunization of Rhesus macaques | | Descriptor: | ANTI-HIV ANTIBODY DH570 FAB HEAVY CHAIN, ANTI-HIV ANTIBODY DH570 FAB HEAVY LIGHT, oligo peptide | | Authors: | Zhang, R, Verkoczy, L, Wiehe, K, Alam, S.M, Nicely, N.I, Santra, S, Bradley, T, Pemble, C, Gao, F, Montefiori, D.C, Bouton-Verville, H, Kelsoe, G, Parks, R, Foulger, A, Tomaras, G, Keple, T.B, Moody, M.A, Liao, H.-X, Haynes, B.F. | | Deposit date: | 2015-08-24 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.488 Å) | | Cite: | Initiation of immune tolerance-controlled HIV gp41 neutralizing B cell lineages.
Sci Transl Med, 8, 2016
|
|
8STZ
 
 | |
5DG1
 
 | | Sugar binding protein - human galectin-2 | | Descriptor: | Galectin-2, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J.Y, Si, Y.L. | | Deposit date: | 2015-08-27 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Human galectin-2 interacts with carbohydrates and peptides non-classically: new insight from X-ray crystallography and hemagglutination.
Acta Biochim.Biophys.Sin., 2016
|
|
5DRR
 
 | | Crystal structure of the Pseudomonas aeruginosa LpxC/LPC-058 complex | | Descriptor: | 4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3S)-4,4-difluoro-3-hydroxy-1-(hydroxyamino)-3-methyl-1-oxobutan-2-yl]benzamide, NITRATE ION, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ... | | Authors: | Lee, C.-J, Najeeb, J, Zhou, P. | | Deposit date: | 2015-09-16 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Drug design from the cryptic inhibitor envelope.
Nat Commun, 7, 2016
|
|
5DSA
 
 | |
5DT9
 
 | | Crystal structure of a putative D-Erythronate-4-Phosphate Dehydrogenase from Vibrio cholerae | | Descriptor: | CHLORIDE ION, Erythronate-4-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Stogios, P.J, Skarina, T, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-09-17 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.663 Å) | | Cite: | Crystal structure of a putative D-Erythronate-4-Phosphate Dehydrogenase from Vibrio cholerae
To Be Published
|
|
8PEM
 
 | |
5DSB
 
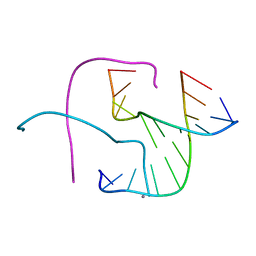 | | Crystal structure of Holliday junctions stabilized by 5-hydroxymethylcytosine in GCC junction core | | Descriptor: | 5'-D(*CP*CP*GP*GP*CP*GP*5HCP*CP*GP*G)-3', CALCIUM ION | | Authors: | Vander Zanden, C.M, Rowe, R.K, Broad, A.J, Ho, P.S. | | Deposit date: | 2015-09-17 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4959 Å) | | Cite: | Effect of Hydroxymethylcytosine on the Structure and Stability of Holliday Junctions.
Biochemistry, 55, 2016
|
|
5DRI
 
 | |
