6VRE
 
 | | Structure of ASK1 bound to BIO-1772961 | | Descriptor: | 3-methoxy-N-{6-[4-(propan-2-yl)-4H-1,2,4-triazol-3-yl]pyridin-2-yl}-1-(pyrazin-2-yl)-1H-pyrazole-4-carboxamide, DIMETHYL SULFOXIDE, Mitogen-activated protein kinase kinase kinase 5 | | Authors: | Chodaparambil, J.V, Marcotte, D.J. | | Deposit date: | 2020-02-07 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Discovery of CNS-Penetrant Apoptosis Signal-Regulating Kinase 1 (ASK1) Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
6VQL
 
 | |
6IP9
 
 | | Crystal Structure of Lanthanum ion (La3+) bound bovine alpha-lactalbumin | | Descriptor: | Alpha-lactalbumin, GLYCEROL, LANTHANUM (III) ION, ... | | Authors: | Prakash, P, Yarramala, S.D, Rao, C.P, Bhaumik, P. | | Deposit date: | 2018-11-02 | | Release date: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Cytotoxicity of apo bovine alpha-lactalbumin complexed with La3+on cancer cells supported by its high resolution crystal structure.
Sci Rep, 9, 2019
|
|
7B3Z
 
 | | Crystal structure of c-MET bound by compound 5 | | Descriptor: | 3-[3-(phenylmethyl)-1~{H}-pyrrolo[2,3-b]pyridin-5-yl]-4,5-dihydro-1~{H}-pyrrolo[3,4-b]pyrrol-6-one, DIMETHYL SULFOXIDE, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
7B3T
 
 | | Crystal structure of c-MET bound by compound 2 | | Descriptor: | 3-(phenylmethyl)-1~{H}-pyrrolo[2,3-b]pyridine, CHLORIDE ION, Hepatocyte growth factor receptor, ... | | Authors: | Collie, G.W. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
7B41
 
 | | Crystal structure of c-MET bound by compound 7 | | Descriptor: | 3-[(2-fluorophenyl)methyl]-5-(1-piperidin-4-ylpyrazol-4-yl)-1~{H}-pyrrolo[2,3-b]pyridine, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2020-12-02 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
7AJR
 
 | | Virtual screening approach leading to the identification of a novel and tractable series of Pseudomonas aeruginosa elastase inhibitors | | Descriptor: | 2-[2-(1,3-benzothiazol-2-ylmethylcarbamoyl)-1,3-dihydroinden-2-yl]ethanoic acid, Keratinase KP2, SULFATE ION, ... | | Authors: | Leiris, S, Davies, D.T, Sprinsky, N, Castandet, J, Behria, L, Bodnarchuk, M.S, Sutton, J.M, Mullins, T.M.G, Jones, M.W, Forrest, A.K, Pallin, T.D, Karunakar, P, Martha, S.K, Parusharamulu, B, Ramula, R, Kotha, V, Pottabathini, N, Pothukanuri, S, Lemonnier, M, Everett, M. | | Deposit date: | 2020-09-29 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Virtual Screening Approach to Identifying a Novel and Tractable Series of Pseudomonas aeruginosa Elastase Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
7B7R
 
 | | MEK1 in complex with compound 4 | | Descriptor: | 2-[5-[ethyl(methyl)amino]imidazo[1,2-a]pyrimidin-7-yl]phenol, Dual specificity mitogen-activated protein kinase kinase 1,Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Kack, H, Oster, L. | | Deposit date: | 2020-12-11 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fragment-Based Discovery of Novel Allosteric MEK1 Binders.
Acs Med.Chem.Lett., 12, 2021
|
|
7B3M
 
 | | MEK1 in complex with compound 6 | | Descriptor: | 1~{H}-indol-2-yl(pyridin-3-yl)methanone, Dual specificity mitogen-activated protein kinase kinase 1,Dual specificity mitogen-activated protein kinase kinase 1, MAGNESIUM ION, ... | | Authors: | Kack, H, Oster, L. | | Deposit date: | 2020-12-01 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fragment-Based Discovery of Novel Allosteric MEK1 Binders.
Acs Med.Chem.Lett., 12, 2021
|
|
5I6N
 
 | | Crystal Structure of Copper Nitrite Reductase at 100K after 11.73 MGy | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRIC OXIDE, ... | | Authors: | Horrell, S, Hough, M.A, Strange, R.W. | | Deposit date: | 2016-02-16 | | Release date: | 2016-07-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Serial crystallography captures enzyme catalysis in copper nitrite reductase at atomic resolution from one crystal.
Iucrj, 3, 2016
|
|
7EZP
 
 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 3-(3-hydroxy-3-oxopropyl)-5-(2-methylpropyl)-7-nitro-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
7EZF
 
 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 7-chloranyl-5-ethyl-3-(3-hydroxy-3-oxopropyl)-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
7EZR
 
 | | Indole-2-carboxylic acid derivatives as allosteric inhibitors of fructose-1,6-bisphosphatase | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 5-ethyl-7-nitro-3-[3-oxidanylidene-3-(thiophen-2-ylsulfonylamino)propyl]-1H-indole-2-carboxylic acid, Fructose-1,6-bisphosphatase 1 | | Authors: | Wang, X.Y, Zhou, J, Xu, B.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Discovery of Novel Indole Derivatives as Fructose-1,6-bisphosphatase Inhibitors and X-ray Cocrystal Structures Analysis.
Acs Med.Chem.Lett., 13, 2022
|
|
5I6K
 
 | |
5I6L
 
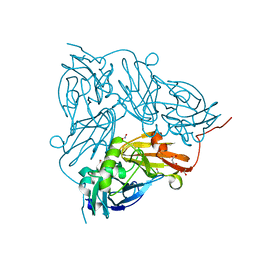 | | Crystal Structure of Copper Nitrite Reductase at 100K after 2.76 MGy | | Descriptor: | ACETATE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Horrell, S, Hough, M.A, Strange, R.W. | | Deposit date: | 2016-02-16 | | Release date: | 2016-07-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Serial crystallography captures enzyme catalysis in copper nitrite reductase at atomic resolution from one crystal.
Iucrj, 3, 2016
|
|
7KBH
 
 | |
1AOE
 
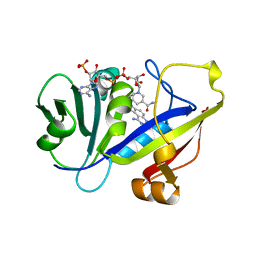 | | CANDIDA ALBICANS DIHYDROFOLATE REDUCTASE COMPLEXED WITH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE (NADPH) AND 1,3-DIAMINO-7-(1-ETHYEPROPYE)-7H-PYRRALO-[3,2-F]QUINAZOLINE (GW345) | | Descriptor: | 7-(1-ETHYL-PROPYL)-7H-PYRROLO-[3,2-F]QUINAZOLINE-1,3-DIAMINE, DIHYDROFOLATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Whitlow, M, Howard, A.J, Stewart, D. | | Deposit date: | 1997-07-02 | | Release date: | 1998-01-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystallographic studies of Candida albicans dihydrofolate reductase. High resolution structures of the holoenzyme and an inhibited ternary complex.
J.Biol.Chem., 272, 1997
|
|
7JYM
 
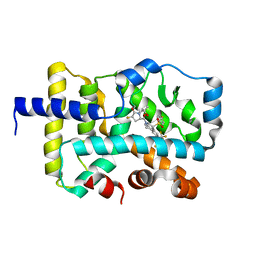 | | CRYSTAL STRUCTURE OF RAR-RELATED ORPHAN RECEPTOR C (NHIS-RORGT(244-487)-L6-SRC1(678-692)) IN COMPLEX WITH A TRICYCLIC SULFONE INVERSE AGONIST | | Descriptor: | (3R,5S)-3-fluoro-5-[(3aR,9bR)-9b-[(4-fluorophenyl)sulfonyl]-7-(1,1,1,2,3,3,3-heptafluoropropan-2-yl)-1,2,3a,4,5,9b-hexahydro-3H-benzo[e]indole-3-carbonyl]-1-(2-hydroxy-2-methylpropyl)pyrrolidin-2-one, Nuclear receptor ROR-gamma, Nuclear receptor coactivator 1 | | Authors: | Sack, J. | | Deposit date: | 2020-08-31 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.051 Å) | | Cite: | Novel Tricyclic Pyroglutamide Derivatives as Potent ROR gamma t Inverse Agonists Identified using a Virtual Screening Approach.
Acs Med.Chem.Lett., 11, 2020
|
|
7K4I
 
 | | Human Arginase 1 in complex with compound 06. | | Descriptor: | 3-[(1~{S},2~{S},5~{R})-2-carboxy-6-thia-3-azabicyclo[3.2.0]heptan-1-yl]propyl-$l^{3}-oxidanyl-bis(oxidanyl)boranuide, Arginase-1, MANGANESE (II) ION | | Authors: | Palte, R.L. | | Deposit date: | 2020-09-15 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Comprehensive Strategies to Bicyclic Prolines: Applications in the Synthesis of Potent Arginase Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
7K4H
 
 | | Human Arginase 1 in complex with compound 04. | | Descriptor: | 3-[(1~{R},5~{S},8~{R})-5-carboxy-2,6-diazabicyclo[3.2.1]octan-8-yl]propyl-$l^{3}-oxidanyl-bis(oxidanyl)boranuide, Arginase-1, MANGANESE (II) ION | | Authors: | Palte, R.L. | | Deposit date: | 2020-09-15 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Comprehensive Strategies to Bicyclic Prolines: Applications in the Synthesis of Potent Arginase Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
7K4J
 
 | | Human Arginase 1 in complex with compound 51. | | Descriptor: | 3-[(1~{S},2~{S},5~{R})-2-carboxy-3,6-diazabicyclo[3.2.0]heptan-1-yl]propyl-$l^{3}-oxidanyl-bis(oxidanyl)boranuide, Arginase-1, MANGANESE (II) ION | | Authors: | Palte, R.L. | | Deposit date: | 2020-09-15 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Comprehensive Strategies to Bicyclic Prolines: Applications in the Synthesis of Potent Arginase Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
7K4G
 
 | | Human Arginase 1 in complex with compound 01. | | Descriptor: | 3-[(2~{S},3~{R})-2-carboxypyrrolidin-3-yl]propyl-$l^{3}-oxidanyl-bis(oxidanyl)boranuide, Arginase-1, MANGANESE (II) ION | | Authors: | Palte, R.L. | | Deposit date: | 2020-09-15 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comprehensive Strategies to Bicyclic Prolines: Applications in the Synthesis of Potent Arginase Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
7K4K
 
 | | Human Arginase 1 in complex with compound 52. | | Descriptor: | 3-[(3~{a}~{S},4~{S},6~{a}~{R})-4-carboxy-2,3,4,5,6,6~{a}-hexahydro-1~{H}-pyrrolo[2,3-c]pyrrol-3~{a}-yl]propyl-$l^{3}-oxidanyl-bis(oxidanyl)boranuide, Arginase-1, MANGANESE (II) ION | | Authors: | Palte, R.L. | | Deposit date: | 2020-09-15 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Comprehensive Strategies to Bicyclic Prolines: Applications in the Synthesis of Potent Arginase Inhibitors.
Acs Med.Chem.Lett., 12, 2021
|
|
1DG3
 
 | | STRUCTURE OF HUMAN GUANYLATE BINDING PROTEIN-1 IN NUCLEOTIDE FREE FORM | | Descriptor: | PROTEIN (INTERFERON-INDUCED GUANYLATE-BINDING PROTEIN 1) | | Authors: | Prakash, B, Praefcke, G.J.K, Renault, L, Wittinghofer, A, Herrmann, C. | | Deposit date: | 1999-11-23 | | Release date: | 2000-10-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human guanylate-binding protein 1 representing a unique class of GTP-binding proteins.
Nature, 403, 2000
|
|
5OF0
 
 | | X-ray structure of human glutamate carboxypeptidase II (GCPII), the E424M inactive mutant, in complex with a inhibitor CFBzOG | | Descriptor: | (2~{S})-2-[[(2~{S})-6-[(4-fluorophenyl)methylamino]-1-oxidanyl-1,6-bis(oxidanylidene)hexan-2-yl]carbamoylamino]pentanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Novakova, Z, Motlova, L, Barinka, C. | | Deposit date: | 2017-07-10 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | 2-Aminoadipic Acid-C(O)-Glutamate Based Prostate-Specific Membrane Antigen Ligands for Potential Use as Theranostics.
ACS Med Chem Lett, 9, 2018
|
|
