8A5X
 
 | |
3E6I
 
 | |
3F49
 
 | |
3GCL
 
 | | Mode of ligand binding and assignment of subsites in mammalian peroxidases: crystal structure of lactoperoxidase complexes with acetyl salycylic acid, salicylhydroxamic acid and benzylhydroxamic acid | | Descriptor: | 2-(ACETYLOXY)BENZOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Singh, A.K, Singh, N, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2009-02-22 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding modes of aromatic ligands to mammalian heme peroxidases with associated functional implications: crystal structures of lactoperoxidase complexes with acetylsalicylic acid, salicylhydroxamic acid, and benzylhydroxamic acid
J.Biol.Chem., 284, 2009
|
|
3DKE
 
 | | Polar and non-polar cavities in phage T4 lysozyme | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AZIDE ION, ... | | Authors: | Liu, L.J, Matthews, B.W. | | Deposit date: | 2008-06-24 | | Release date: | 2008-11-25 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Use of experimental crystallographic phases to examine the hydration of polar and nonpolar cavities in T4 lysozyme
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3FO5
 
 | | Human START domain of Acyl-coenzyme A thioesterase 11 (ACOT11) | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Siponen, M.I, Lehtio, L, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Thorsell, A.G, Tresaugues, L, Van-Den-Berg, S, Weigelt, J, Welin, M, Wikstrom, M, Wisniewska, M, Shueler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-27 | | Release date: | 2009-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Comparative structural analysis of lipid binding START domains.
Plos One, 6, 2011
|
|
3FU7
 
 | | Melanocarpus albomyces laccase crystal soaked (4 sec) with 2,6-dimethoxyphenol | | Descriptor: | 2,6-dimethoxycyclohexa-2,5-diene-1,4-dione, 2,6-dimethoxyphenol, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kallio, J.P, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2009-01-14 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure Function Studies of a Melanocarpus albomyces Laccase Suggest a Pathway for Oxidation of Phenolic Compounds.
J.Mol.Biol., 392, 2009
|
|
3GFR
 
 | | Structure of YhdA, D137L variant | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADPH-azoreductase | | Authors: | Staunig, N, Gruber, K. | | Deposit date: | 2009-02-27 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | A single intersubunit salt bridge affects oligomerization and catalytic activity in a bacterial quinone reductase
Febs J., 276, 2009
|
|
3FYG
 
 | | CRYSTAL STRUCTURE OF TETRADECA-(3-FLUOROTYROSYL)-GLUTATHIONE S-TRANSFERASE | | Descriptor: | (9R,10R)-9-(S-GLUTATHIONYL)-10-HYDROXY-9,10-DIHYDROPHENANTHRENE, MU CLASS TETRADECA-(3-FLUOROTYROSYL)-GLUTATHIONE S-TRANSFERASE OF ISOENZYME | | Authors: | Xiao, G, Parsons, J.F, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1997-08-07 | | Release date: | 1999-06-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational changes in the crystal structure of rat glutathione transferase M1-1 with global substitution of 3-fluorotyrosine for tyrosine.
J.Mol.Biol., 281, 1998
|
|
3FU8
 
 | | Melanocarpus albomyces laccase crystal soaked (10 sec) with 2,6-dimethoxyphenol | | Descriptor: | 2,6-dimethoxyphenol, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kallio, J.P, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2009-01-14 | | Release date: | 2009-09-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Function Studies of a Melanocarpus albomyces Laccase Suggest a Pathway for Oxidation of Phenolic Compounds.
J.Mol.Biol., 392, 2009
|
|
3GFQ
 
 | | Structure of YhdA, K109L variant | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADPH-azoreductase | | Authors: | Staunig, N, Gruber, K. | | Deposit date: | 2009-02-27 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.996 Å) | | Cite: | A single intersubunit salt bridge affects oligomerization and catalytic activity in a bacterial quinone reductase
Febs J., 276, 2009
|
|
3GFS
 
 | | Structure of YhdA, K109D/D137K variant | | Descriptor: | FLAVIN MONONUCLEOTIDE, FMN-dependent NADPH-azoreductase | | Authors: | Staunig, N, Gruber, K. | | Deposit date: | 2009-02-27 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | A single intersubunit salt bridge affects oligomerization and catalytic activity in a bacterial quinone reductase
Febs J., 276, 2009
|
|
3FF0
 
 | |
3GUR
 
 | | Crystal Structure of mu class glutathione S-transferase (GSTM2-2) in complex with glutathione and 6-(7-Nitro-2,1,3-benzoxadiazol-4-ylthio)hexanol (NBDHEX) | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, Glutathione S-transferase Mu 2, ... | | Authors: | Federici, L, Lo Sterzo, C, Di Matteo, A, Scaloni, F, Federici, G, Caccuri, A.M. | | Deposit date: | 2009-03-30 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the binding of the anticancer compound 6-(7-nitro-2,1,3-benzoxadiazol-4-ylthio)hexanol to human glutathione s-transferases
Cancer Res., 69, 2009
|
|
3H4K
 
 | | Crystal structure of the wild type Thioredoxin glutatione reductase from Schistosoma mansoni in complex with auranofin | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE, GOLD ION, ... | | Authors: | Angelucci, F, Dimastrogiovanni, D, Miele, A.E, Boumis, G, Brunori, M, Bellelli, A. | | Deposit date: | 2009-04-20 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Inhibition of Schistosoma mansoni thioredoxin-glutathione reductase by auranofin: structural and kinetic aspects.
J.Biol.Chem., 284, 2009
|
|
3DKH
 
 | | L559A mutant of Melanocarpus albomyces laccase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hakulinen, N, Rouvinen, J. | | Deposit date: | 2008-06-25 | | Release date: | 2009-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Essential role of the C-terminus in Melanocarpus albomyces laccase for enzyme production, catalytic properties and structure
Febs J., 276, 2009
|
|
3GPH
 
 | |
3E4E
 
 | |
3GST
 
 | | STRUCTURE OF THE XENOBIOTIC SUBSTRATE BINDING SITE OF A GLUTATHIONE S-TRANSFERASE AS REVEALED BY X-RAY CRYSTALLOGRAPHIC ANALYSIS OF PRODUCT COMPLEXES WITH THE DIASTEREOMERS OF 9-(S-GLUTATHIONYL)-10-HYDROXY-9, 10-DIHYDROPHENANTHRENE | | Descriptor: | (9R,10R)-9-(S-GLUTATHIONYL)-10-HYDROXY-9,10-DIHYDROPHENANTHRENE, GLUTATHIONE S-TRANSFERASE, SULFATE ION | | Authors: | Ji, X, Ammon, H.L, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1993-06-07 | | Release date: | 1993-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of the xenobiotic substrate binding site of a glutathione S-transferase as revealed by X-ray crystallographic analysis of product complexes with the diastereomers of 9-(S-glutathionyl)-10-hydroxy-9,10-dihydrophenanthrene.
Biochemistry, 33, 1994
|
|
3GWR
 
 | |
3H6K
 
 | |
3GTU
 
 | |
3HIG
 
 | |
3HFG
 
 | |
5OQM
 
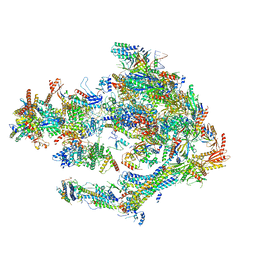 | | STRUCTURE OF YEAST TRANSCRIPTION PRE-INITIATION COMPLEX WITH TFIIH AND CORE MEDIATOR | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Schilbach, S, Hantsche, M, Tegunov, D, Dienemann, C, Wigge, C, Urlaub, H, Cramer, P. | | Deposit date: | 2017-08-13 | | Release date: | 2018-05-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structures of transcription pre-initiation complex with TFIIH and Mediator.
Nature, 551, 2017
|
|
