4YNU
 
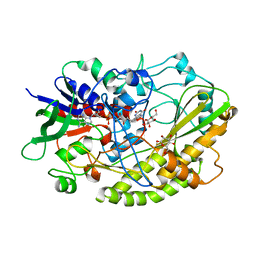 | | Crystal structure of Aspergillus flavus FADGDH in complex with D-glucono-1,5-lactone | | Descriptor: | D-glucono-1,5-lactone, FLAVIN-ADENINE DINUCLEOTIDE, Glucose oxidase, ... | | Authors: | Yoshida, H, Sakai, G, Kojima, K, Kamitori, S, Sode, K. | | Deposit date: | 2015-03-11 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural analysis of fungus-derived FAD glucose dehydrogenase
Sci Rep, 5, 2015
|
|
8BGR
 
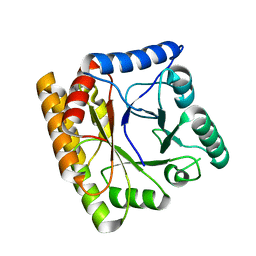 | |
2AXR
 
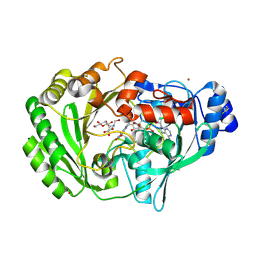 | | Crystal structure of glucooligosaccharide oxidase from Acremonium strictum: a novel flavinylation of 6-S-cysteinyl, 8alpha-N1-histidyl FAD | | Descriptor: | (2R,3R,4R,5R)-4,5-dihydroxy-2-(hydroxymethyl)-6-oxopiperidin-3-yl beta-D-glucopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Huang, C.-H, Lai, W.-L, Vasella, A, Tsai, Y.-C, Liaw, S.-H. | | Deposit date: | 2005-09-05 | | Release date: | 2005-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of Glucooligosaccharide Oxidase from Acremonium strictum: A NOVEL FLAVINYLATION OF 6-S-CYSTEINYL, 8{alpha}-N1-HISTIDYL FAD
J.Biol.Chem., 280, 2005
|
|
1AMO
 
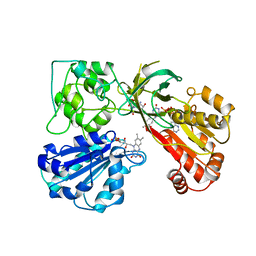 | | THREE-DIMENSIONAL STRUCTURE OF NADPH-CYTOCHROME P450 REDUCTASE: PROTOTYPE FOR FMN-AND FAD-CONTAINING ENZYMES | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Wang, M, Roberts, D.L, Paschke, R, Shea, T.M, Masters, B.S.S, Kim, J.J.P. | | Deposit date: | 1997-06-17 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Three-dimensional structure of NADPH-cytochrome P450 reductase: prototype for FMN- and FAD-containing enzymes.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
2E0I
 
 | | Crystal structure of archaeal photolyase from Sulfolobus tokodaii with two FAD molecules: Implication of a novel light-harvesting cofactor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 432aa long hypothetical deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fujihashi, M, Numoto, N, Kobayashi, Y, Mizushima, A, Tsujimura, M, Nakamura, A, Kawarabayashi, Y, Miki, K. | | Deposit date: | 2006-10-10 | | Release date: | 2006-11-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Archaeal Photolyase from Sulfolobus tokodaii with Two FAD Molecules: Implication of a Novel Light-harvesting Cofactor
J.Mol.Biol., 365, 2007
|
|
4B4D
 
 | | Crystal structure of FAD-containing ferredoxin-NADP reductase from Xanthomonas axonopodis pv. citri | | Descriptor: | CHLORIDE ION, FERREDOXIN-NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Martinez-Julvez, M, Orellano, E.G, Tondo, M.L, Hurtado-Guerrero, R, Medina, M, Ceccarelli, E.A, Sanchez-Azqueta, A. | | Deposit date: | 2012-07-30 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Fad-Containing Ferredoxin- Nadp Reductase from Xanthomonas Axonopodis Pv. Citri
Biomed Res Int, 2013, 2013
|
|
3E1T
 
 | | Structure and action of the myxobacterial chondrochloren halogenase CndH, a new variant of FAD-dependent halogenases | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Halogenase | | Authors: | Buedenbender, S, Rachid, S, Mueller, R, Schulz, G.E. | | Deposit date: | 2008-08-04 | | Release date: | 2009-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and action of the myxobacterial chondrochloren halogenase CndH: a new variant of FAD-dependent halogenases.
J.Mol.Biol., 385, 2009
|
|
1F20
 
 | | CRYSTAL STRUCTURE OF RAT NEURONAL NITRIC-OXIDE SYNTHASE FAD/NADP+ DOMAIN AT 1.9A RESOLUTION. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, GLYCEROL, ... | | Authors: | Zhang, J, Martasek, P, Masters, B.S, Kim, J.P. | | Deposit date: | 2000-05-22 | | Release date: | 2001-10-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the FAD/NADPH-binding domain of rat neuronal nitric-oxide synthase. Comparisons with NADPH-cytochrome P450 oxidoreductase.
J.Biol.Chem., 276, 2001
|
|
6A2U
 
 | | Crystal structure of gamma-alpha subunit complex from Burkholderia cepacia FAD glucose dehydrogenase | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Glucose dehydrogenase, ... | | Authors: | Yoshida, H, Kojima, K, Yoshimatsu, K, Shiota, M, Yamazaki, T, Ferri, S, Tsugawa, W, Kamitori, S, Sode, K. | | Deposit date: | 2018-06-13 | | Release date: | 2019-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-ray structure of the direct electron transfer-type FAD glucose dehydrogenase catalytic subunit complexed with a hitchhiker protein.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
4IFX
 
 | | Crystal structure of Treponema pallidum TP0796 Flavin trafficking protein, FAD substrate bound form | | Descriptor: | ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2012-12-15 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.452 Å) | | Cite: | The TP0796 Lipoprotein of Treponema pallidum Is a Bimetal-dependent FAD Pyrophosphatase with a Potential Role in Flavin Homeostasis.
J.Biol.Chem., 288, 2013
|
|
4DQL
 
 | |
3MNR
 
 | | Crystal Structure of Benzamide SNX-1321 bound to Hsp90 | | Descriptor: | 2-[(3,4,5-trimethoxyphenyl)amino]-4-(2,6,6-trimethyl-4-oxo-4,5,6,7-tetrahydro-1H-indol-1-yl)benzamide, Heat shock protein HSP 90-alpha | | Authors: | Veal, J.M, Fadden, P, Huang, K.H, Rice, J, Hall, S.E, Haytstead, T.A. | | Deposit date: | 2010-04-22 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Application of Chemoproteomics to Drug Discovery: Identification of a Clinical Candidate Targeting Hsp90.
Chem.Biol., 17, 2010
|
|
4P96
 
 | |
8HCZ
 
 | |
8HDF
 
 | | Full length crystal structure of mycobacterium tuberculosis FadD23 in complex with ANP and PLM | | Descriptor: | Long-chain-fatty-acid--AMP ligase FadD23, PALMITIC ACID, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yan, M.R, Liu, X, Zhang, W, Rao, Z.H. | | Deposit date: | 2022-11-04 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The Key Roles of Mycobacterium tuberculosis FadD23 C-terminal Domain in Catalytic Mechanisms.
Front Microbiol, 14, 2023
|
|
8HD4
 
 | | Full-length crystal structure of mycobacterium tuberculosis FadD23 in complex with AMPC16 | | Descriptor: | Long-chain-fatty-acid--AMP ligase FadD23, palmitoyl adenylate | | Authors: | Yan, M.R, Liu, X, Zhang, W, Rao, Z.H. | | Deposit date: | 2022-11-03 | | Release date: | 2023-02-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | The Key Roles of Mycobacterium tuberculosis FadD23 C-terminal Domain in Catalytic Mechanisms.
Front Microbiol, 14, 2023
|
|
8IQU
 
 | | Structure of MtbFadD23 with PhU-AMS | | Descriptor: | 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine, Fatty-acid-CoA ligase FadD23 | | Authors: | Yan, M.R, Zhang, W. | | Deposit date: | 2023-03-17 | | Release date: | 2023-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural basis for the development of potential inhibitors targeting FadD23 from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
7XLY
 
 | | Crystal structure of FadA2 (Rv0243) from the fatty acid metabolic pathway of Mycobacterium tuberculosis | | Descriptor: | Probable acetyl-CoA acyltransferase FadA2 (3-ketoacyl-CoA thiolase) (Beta-ketothiolase), SULFATE ION | | Authors: | Singh, R, Kundu, P, Singh, B.K, Bhattacharyya, S, Das, A.K. | | Deposit date: | 2022-04-23 | | Release date: | 2023-04-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of FadA2 thiolase from Mycobacterium tuberculosis and prediction of its substrate specificity and membrane-anchoring properties.
Febs J., 290, 2023
|
|
5HM3
 
 | | 2.25 Angstrom Resolution Crystal Structure of Long-chain-fatty-acid-AMP Ligase FadD32 from Mycobacterium tuberculosis in complex with Inhibitor 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine | | Descriptor: | 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Warwrzak, Z, Kuhn, M.L, Shuvalova, L, Flores, K.J, Wilson, D.J, Grimes, K.D, Aldrich, C.C, Anderson, W.A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-15 | | Release date: | 2016-08-03 | | Last modified: | 2016-09-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the Essential Mtb FadD32 Enzyme: A Promising Drug Target for Treating Tuberculosis.
Acs Infect Dis., 2, 2016
|
|
5EY9
 
 | | Structure of FadD32 from Mycobacterium marinum complexed to AMPC12 | | Descriptor: | GLYCEROL, Long-chain-fatty-acid--AMP ligase FadD32, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl icosyl hydrogen phosphate | | Authors: | Guillet, V, Maveyraud, L, Mourey, L. | | Deposit date: | 2015-11-24 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight into Structure-Function Relationships and Inhibition of the Fatty Acyl-AMP Ligase (FadD32) Orthologs from Mycobacteria.
J.Biol.Chem., 291, 2016
|
|
6AMB
 
 | | Crystal Structure of the Afadin RA1 domain in complex with HRAS | | Descriptor: | Afadin, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Smith, M.J, Ishiyama, N, Ikura, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evolution of AF6-RAS association and its implications in mixed-lineage leukemia.
Nat Commun, 8, 2017
|
|
6M7X
 
 | | Structure of human CYP11B1 in complex with fadrozole | | Descriptor: | 4-[(5S)-5,6,7,8-tetrahydroimidazo[1,5-a]pyridin-5-yl]benzonitrile, Cytochrome P450 11B1, mitochondrial, ... | | Authors: | Scott, E.E, Brixius-Anderko, S. | | Deposit date: | 2018-08-21 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Structure of human cortisol-producing cytochrome P450 11B1 bound to the breast cancer drug fadrozole provides insights for drug design.
J. Biol. Chem., 294, 2019
|
|
4ISB
 
 | |
4IR7
 
 | | Crystal Structure of Mtb FadD10 in Complex with Dodecanoyl-AMP | | Descriptor: | 5'-O-[(S)-(dodecanoyloxy)(hydroxy)phosphoryl]adenosine, Long chain fatty acid CoA ligase FadD10, MAGNESIUM ION | | Authors: | Liu, Z, Wang, F, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2013-01-14 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Mycobacterium tuberculosis FadD10 protein reveal a new type of adenylate-forming enzyme.
J.Biol.Chem., 288, 2013
|
|
7LO1
 
 | |
