3CVS
 
 | | Crystal Structure of an AlkA Host/Guest Complex 8oxoGuanine:Adenine Base Pair | | Descriptor: | DNA (5'-D(*DGP*DAP*DCP*DAP*DTP*DGP*DAP*(8OG)P*DTP*DGP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DCP*DAP*DAP*DTP*DCP*DAP*DTP*DGP*DTP*DC)-3'), DNA-3-methyladenine glycosylase 2 | | Authors: | Bowman, B.R, Lee, S, Wang, S, Verdine, G.L. | | Deposit date: | 2008-04-19 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the E. coli DNA Glycosylase AlkA Bound to the Ends of Duplex DNA: A System for the Structure Determination of Lesion-Containing DNA.
Structure, 16, 2008
|
|
3CYJ
 
 | | Crystal structure of a mandelate racemase/muconate lactonizing enzyme-like protein from Rubrobacter xylanophilus | | Descriptor: | GLYCEROL, Mandelate racemase/muconate lactonizing enzyme-like protein, SODIUM ION | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Zhang, F, Bravo, J, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-25 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a mandelate racemase/muconate lactonizing enzyme-like protein from Rubrobacter xylanophilus.
To be Published
|
|
3LMF
 
 | | Crystal Structure of Nmul_A1745 protein from Nitrosospira multiformis, Northeast Structural Genomics Consortium Target NmR72 | | Descriptor: | Uncharacterized protein | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-29 | | Release date: | 2010-02-16 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Northeast Structural Genomics Consortium Target NmR72
To be Published
|
|
3LOO
 
 | | Crystal structure of Anopheles gambiae adenosine kinase in complex with P1,P4-di(adenosine-5) tetraphosphate | | Descriptor: | Anopheles gambiae adenosine kinase, BIS(ADENOSINE)-5'-TETRAPHOSPHATE, CHLORIDE ION, ... | | Authors: | Ho, M.-C, Cassera, M.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2010-02-04 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A High-Affinity Adenosine Kinase from Anopheles gambiae.
Biochemistry, 50, 2011
|
|
3LW2
 
 | | Mouse Plasminogen Activator Inhibitor-1 (PAI-1) | | Descriptor: | Plasminogen activator inhibitor 1 | | Authors: | Dewilde, M, Van De Craen, B, Compernolle, G, Madsen, J.B, Strelkov, S.V, Gils, A, Declerck, P.J. | | Deposit date: | 2010-02-23 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Subtle structural differences between human and mouse PAI-1 reveal the basis for biochemical differences.
J.Struct.Biol., 171, 2010
|
|
4UCE
 
 | | N-terminal globular domain of the RSV Nucleoprotein in complex with the Nucleoprotein Phosphoprotein interaction inhibitor M72 | | Descriptor: | 1-[(4-fluorophenyl)methyl]pyrazole-3,5-dicarboxylic acid, NUCLEOPROTEIN, SULFATE ION | | Authors: | Ouizougun-Oubari, M, Pereira, N, Tarus, B, Galloux, M, Tortorici, M.-A, Hoos, S, Baron, B, England, P, Bontems, F, Rey, F.A, Eleouet, J.-F, Sizun, C, Slama-Schwok, A, Duquerroy, S. | | Deposit date: | 2014-12-03 | | Release date: | 2015-08-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | A Druggable Pocket at the Nucleocapsid/Phosphoprotein Interaction Site of the Human Respiratory Syncytial Virus.
J.Virol., 89, 2015
|
|
4UC7
 
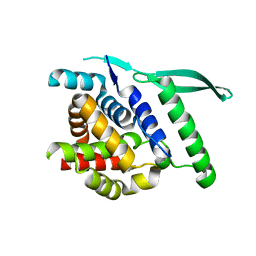 | | N-terminal globular domain of the RSV Nucleoprotein | | Descriptor: | NUCLEOPROTEIN, SULFATE ION | | Authors: | Ouizougun-Oubari, M, Pereira, N, Tarus, B, Galloux, M, Tortorici, M.-A, Hoos, S, Baron, B, England, P, Bontems, F, Rey, F.A, Eleouet, J.-F, Sizun, C, Slama-Schwok, A, Duquerroy, S. | | Deposit date: | 2014-12-03 | | Release date: | 2015-08-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A Druggable Pocket at the Nucleocapsid/Phosphoprotein Interaction Site of the Human Respiratory Syncytial Virus.
J.Virol., 89, 2015
|
|
4UCB
 
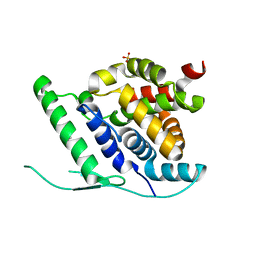 | | N-terminal globular domain of the RSV Nucleoprotein in complex with C- terminal peptide of the Phosphoprotein | | Descriptor: | NUCLEOPROTEIN, PHOSPHOSPROTEIN, SULFATE ION | | Authors: | Ouizougun-Oubari, M, Pereira, N, Tarus, B, Galloux, M, Tortorici, M.-A, Hoos, S, Baron, B, England, P, Bontems, F, Rey, F.A, Eleouet, J.-F, Sizun, C, Slama-Schwok, A, Duquerroy, S. | | Deposit date: | 2014-12-03 | | Release date: | 2015-08-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | A Druggable Pocket at the Nucleocapsid/Phosphoprotein Interaction Site of the Human Respiratory Syncytial Virus.
J.Virol., 89, 2015
|
|
3CWA
 
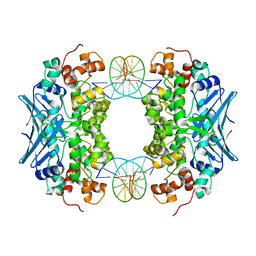 | | Crystal Structure of an AlkA Host/Guest Complex 8oxoGuanine:Cytosine Base Pair | | Descriptor: | DNA (5'-D(*(8OG)P*DAP*DCP*DAP*DTP*DGP*DAP*DGP*DTP*DGP*DCP*DC)-3'), DNA (5'-D(*DGP*DGP*DCP*DAP*DCP*DTP*DCP*DAP*DTP*DGP*DTP*DC)-3'), DNA-3-methyladenine glycosylase 2 | | Authors: | Bowman, B.R, Lee, S, Wang, S, Verdine, G.L. | | Deposit date: | 2008-04-21 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the E. coli DNA Glycosylase AlkA Bound to the Ends of Duplex DNA: A System for the Structure Determination of Lesion-Containing DNA.
Structure, 16, 2008
|
|
3D4V
 
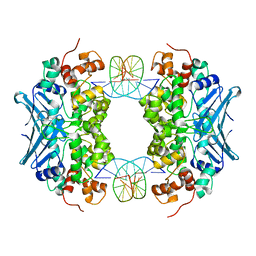 | | Crystal Structure of an AlkA Host/Guest Complex N7MethylGuanine:Cytosine Base Pair | | Descriptor: | 5'-D(*DGP*DAP*DCP*DAP*DTP*DGP*DAP*(FMG)P*DTP*DGP*DCP*DC)-3', 5'-D(*DGP*DGP*DCP*DAP*DCP*DTP*DCP*DAP*DTP*DGP*DTP*DC)-3', DNA-3-methyladenine glycosylase 2 | | Authors: | Lee, S, Bowman, B.R, Wang, S, Verdine, G.L. | | Deposit date: | 2008-05-15 | | Release date: | 2008-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Synthesis and structure of duplex DNA containing the genotoxic nucleobase lesion N7-methylguanine.
J.Am.Chem.Soc., 130, 2008
|
|
3LUC
 
 | |
3LUI
 
 | | Crystal structure of the SNX17 PX domain with bound sulphate | | Descriptor: | SULFATE ION, Sorting nexin-17 | | Authors: | Ghai, R, Collins, B.M. | | Deposit date: | 2010-02-17 | | Release date: | 2010-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Phox homology band 4.1/ezrin/radixin/moesin-like proteins function as molecular scaffolds that interact with cargo receptors and Ras GTPases
Proc.Natl.Acad.Sci.USA, 2011
|
|
3D5S
 
 | | Crystal Structure of Efb-C (R131A) / C3d Complex | | Descriptor: | Complement C3, Fibrinogen-binding protein | | Authors: | Geisbrecht, B.V. | | Deposit date: | 2008-05-16 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Electrostatic contributions drive the interaction between Staphylococcus aureus protein Efb-C and its complement target C3d.
Protein Sci., 17, 2008
|
|
3LX4
 
 | | Stepwise [FeFe]-hydrogenase H-cluster assembly revealed in the structure of HydA(deltaEFG) | | Descriptor: | ACETATE ION, CHLORIDE ION, Fe-hydrogenase, ... | | Authors: | Mulder, D.W, Boyd, E.S, Sarma, R, Lange, R.K, Endrizzi, J.A, Broderick, J.B, Peters, J.W. | | Deposit date: | 2010-02-24 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Stepwise [FeFe]-hydrogenase H-cluster assembly revealed in the structure of HydA(DeltaEFG).
Nature, 465, 2010
|
|
4UOS
 
 | | Thermodynamic hyperstability in parametrically designed helical bundles | | Descriptor: | DESIGNED HELICAL BUNDLE | | Authors: | Oberdorfer, G, Huang, P, Pei, X.Y, Xu, C, Gonen, T, Nannenga, B, DiMaio, D, Rogers, J, Luisi, B.F, Baker, D. | | Deposit date: | 2014-06-09 | | Release date: | 2014-11-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | High Thermodynamic Stability of Parametrically Designed Helical Bundles
Science, 346, 2014
|
|
4V2R
 
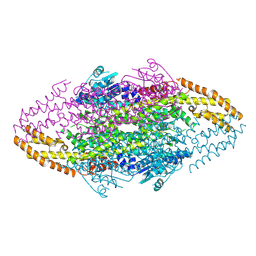 | | Ironing out their differences: Dissecting the structural determinants of a phenylalanine aminomutase and ammonia lyase | | Descriptor: | PHENYLALANINE AMINOMUTASE (L-BETA-PHENYLALANINE FORMING) | | Authors: | Heberling, M, Masman, M, Bartsch, S, Wybenga, G.G, Dijkstra, B.W, Marrink, S, Janssen, D. | | Deposit date: | 2014-10-14 | | Release date: | 2014-12-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ironing out their differences: dissecting the structural determinants of a phenylalanine aminomutase and ammonia lyase.
ACS Chem. Biol., 10, 2015
|
|
7TBJ
 
 | | Composite structure of the human nuclear pore complex (NPC) symmetric core generated with a 12A cryo-ET map of the purified HeLa cell NPC | | Descriptor: | NUP107 CTD, NUP107 NTD, NUP133, ... | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-12-22 | | Release date: | 2022-06-22 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
7TBL
 
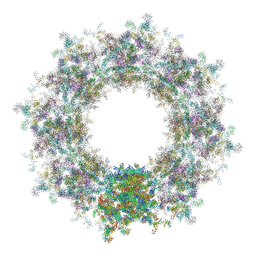 | | Composite structure of the human nuclear pore complex (NPC) cytoplasmic face generated with a 12A cryo-ET map of the purified HeLa cell NPC | | Descriptor: | DDX19, ELYS, GLE1, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-12-22 | | Release date: | 2022-06-22 | | Last modified: | 2022-06-29 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7TC6
 
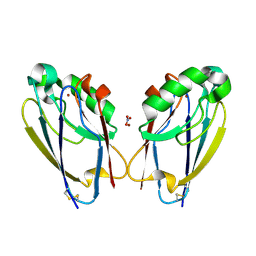 | | All Phe-Azurin variant - F15W | | Descriptor: | Azurin, COPPER (II) ION, NITRATE ION | | Authors: | Fedoretz-Maxwell, B.P, Worrall, L.J, Strynadka, N.C.J, Warren, J.J. | | Deposit date: | 2021-12-22 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Impact of Second Coordination Sphere Methionine-Aromatic Interactions in Copper Proteins.
Inorg.Chem., 61, 2022
|
|
3MAP
 
 | |
3MBF
 
 | |
7TF7
 
 | | S. aureus GS(12) - apo | | Descriptor: | Glutamine synthetase | | Authors: | Travis, B.A, Peck, J, Schumacher, M.A. | | Deposit date: | 2022-01-06 | | Release date: | 2022-06-29 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.13 Å) | | Cite: | Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria.
Nat Commun, 13, 2022
|
|
4UZF
 
 | |
3M41
 
 | | Crystal structure of the mutant V182A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum | | Descriptor: | GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2010-03-10 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
3ME4
 
 | | Crystal structure of mouse RANK | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Walter, S.W, Liu, C, Zhu, X, Wu, Y, Owens, R.J, Stuart, D.I, Gao, B, Ren, J. | | Deposit date: | 2010-03-31 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and Functional Insights of RANKL-RANK Interaction and Signaling.
J.Immunol., 2010
|
|
