2MNR
 
 | |
3CT2
 
 | | Crystal structure of muconate cycloisomerase from Pseudomonas fluorescens | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-11 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: stereochemically distinct mechanisms in two families of cis,cis-muconate lactonizing enzymes.
Biochemistry, 48, 2009
|
|
1MUC
 
 | | STRUCTURE OF MUCONATE LACTONIZING ENZYME AT 1.85 ANGSTROMS RESOLUTION | | Descriptor: | MANGANESE (II) ION, MUCONATE LACTONIZING ENZYME | | Authors: | Helin, S, Kahn, P.C, Guha, B.H.L, Mallows, D.J, Goldman, A. | | Deposit date: | 1995-09-20 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The refined X-ray structure of muconate lactonizing enzyme from Pseudomonas putida PRS2000 at 1.85 A resolution.
J.Mol.Biol., 254, 1995
|
|
3FJ4
 
 | | Crystal structure of muconate lactonizing enzyme from Pseudomonas Fluorescens complexed with muconolactone | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase, [(2S)-5-oxo-2,5-dihydrofuran-2-yl]acetic acid | | Authors: | Fedorov, A.A, Fedorov, E.V, Sakai, A, Gerlt, J.A, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-12-14 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: stereochemically distinct mechanisms in two families of cis,cis-muconate lactonizing enzymes
Biochemistry, 48, 2009
|
|
3DGB
 
 | | Crystal structure of muconate lactonizing enzyme from Pseudomonas Fluorescens complexed with muconolactone | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase, [(2S)-5-oxo-2,5-dihydrofuran-2-yl]acetic acid | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-13 | | Release date: | 2009-03-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: stereochemically distinct mechanisms in two families of cis,cis-muconate lactonizing enzymes
Biochemistry, 48, 2009
|
|
8KC9
 
 | | Human collagen prolyl processing enzyme complex, P3H1/CRTAP/PPIB heterotrimer, bound to cyclosporin A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cartilage-associated protein, DSN-MLE-MLE-MVA-BMT-ABA-SAR-MLE-VAL-MLE-ALA, ... | | Authors: | Li, W, Peng, J, Yao, D, Rao, B, Xia, Y, Wang, Q, Li, S, Cao, M, Shen, Y, Ma, P, Liao, R, Qin, A, Zhao, J, Cao, Y. | | Deposit date: | 2023-08-06 | | Release date: | 2024-09-18 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | The structural basis for the collagen processing by human P3H1/CRTAP/PPIB ternary complex.
Nat Commun, 15, 2024
|
|
8CUN
 
 | |
8CWA
 
 | |
4WC8
 
 | |
8JJS
 
 | | Human K-Ras G12D (GDP-bound) in complex with cyclic peptide inhibitor AP10343 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, MAA-ILE-SAR-SAR-7T2-SAR-IAE-LEU-MEA-MLE-7TK, ... | | Authors: | Irie, M, Fukami, T.A, Tanada, M, Ohta, A, Torizawa, T. | | Deposit date: | 2023-05-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | Development of Orally Bioavailable Peptides Targeting an Intracellular Protein: From a Hit to a Clinical KRAS Inhibitor.
J.Am.Chem.Soc., 145, 2023
|
|
7UBF
 
 | |
7UBI
 
 | |
7UBE
 
 | |
8SZQ
 
 | | Cat DHX9 bound to ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Lee, Y.-T, Sickmier, E.A, Grigoriu, S, Boriack-Sjodin, P.A. | | Deposit date: | 2023-05-30 | | Release date: | 2023-08-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.711 Å) | | Cite: | Crystal structures of the DExH-box RNA helicase DHX9.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8SZP
 
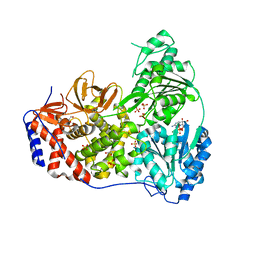 | | Human DHX9 bound to ADP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase A, ... | | Authors: | Lee, Y.-T, Sickmier, E.A, Grigoriu, S, Boriack-Sjodin, P.A. | | Deposit date: | 2023-05-30 | | Release date: | 2023-08-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal structures of the DExH-box RNA helicase DHX9.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8SZR
 
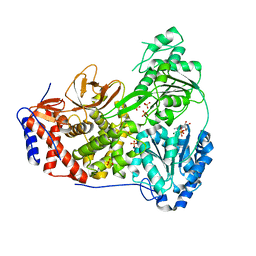 | | Dog DHX9 bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RNA helicase, ... | | Authors: | Lee, Y.-T, Sickmier, E.A, Grigoriu, S, Boriack-Sjodin, P.A. | | Deposit date: | 2023-05-30 | | Release date: | 2023-08-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Crystal structures of the DExH-box RNA helicase DHX9.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7JRH
 
 | |
5W5U
 
 | |
5W6T
 
 | |
7UZL
 
 | |
7UBC
 
 | |
7UBG
 
 | |
3VEU
 
 | | Crystal Structure of Human Beta Secretase in Complex with NVP-AVI326 | | Descriptor: | (2S)-N-[(2S,3R)-3-hydroxy-1-phenyl-4-{[3-(propan-2-yl)benzyl]amino}butan-2-yl]-2-[(5S)-6-oxo-1-propyl-1,7-diazaspiro[4.4]non-7-yl]propanamide, Beta-secretase 1 | | Authors: | Rondeau, J.M, Bourgier, E. | | Deposit date: | 2012-01-09 | | Release date: | 2012-11-21 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery of cyclic sulfone hydroxyethylamines as potent and selective beta-site APP-cleaving enzyme 1 (BACE1) inhibitors: structure based design and in vivo reduction of amyloid beta-peptides
J.Med.Chem., 55, 2012
|
|
1O1E
 
 | | MOLECULAR MODELS OF AVERAGED RIGOR CROSSBRIDGES FROM TOMOGRAMS OF INSECT FLIGHT MUSCLE | | Descriptor: | SKELETAL MUSCLE ACTIN, SKELETAL MUSCLE MYOSIN II, SKELETAL MUSCLE MYOSIN II ESSENTIAL LIGHT CHAIN, ... | | Authors: | Chen, L.F, Winkler, H, Reedy, M.K, Reedy, M.C, Taylor, K.A. | | Deposit date: | 2002-11-19 | | Release date: | 2002-12-04 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (70 Å) | | Cite: | Molecular Modeling of Averaged Rigor Crossbridges from Tomograms of Insect Flight Muscle
J.Struct.Biol., 138, 2002
|
|
1O19
 
 | | MOLECULAR MODELS OF AVERAGED RIGOR CROSSBRIDGES FROM TOMOGRAMS OF INSECT FLIGHT MUSCLE | | Descriptor: | SKELETAL MUSCLE ACTIN, SKELETAL MUSCLE MYOSIN II, SKELETAL MUSCLE MYOSIN II ESSENTIAL LIGHT CHAIN, ... | | Authors: | Chen, L.F, Winkler, H, Reedy, M.K, Reedy, M.C, Taylor, K.A. | | Deposit date: | 2002-11-15 | | Release date: | 2002-11-27 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (70 Å) | | Cite: | Molecular Modeling of Averaged Rigor Crossbridges from Tomograms of Insect Flight Muscle
J.Struct.Biol., 138, 2002
|
|
