3OVZ
 
 | | Cathepsin K in complex with a covalent inhibitor with a ketoamide warhead | | Descriptor: | Cathepsin K, N-[(1S)-3-amino-1-ethyl-2,3-dioxopropyl]-2-chloro-4-(pyridin-2-ylmethoxy)-3-(trifluoromethyl)benzamide, SULFATE ION | | Authors: | Fradera, X, van Zeeland, M, Uitdehaag, J.C.M. | | Deposit date: | 2010-09-17 | | Release date: | 2010-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Trifluoromethylphenyl as P2 for ketoamide-based cathepsin S inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3P0T
 
 | |
4DVH
 
 | | Crystal structure of Trypanosoma cruzi mitochondrial iron superoxide dismutase | | Descriptor: | FE (III) ION, Superoxide dismutase | | Authors: | Larrieux, N, Buschiazzo, A. | | Deposit date: | 2012-02-23 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural and Molecular Basis of the Peroxynitrite-mediated Nitration and Inactivation of Trypanosoma cruzi Iron-Superoxide Dismutases (Fe-SODs) A and B: DISPARATE SUSCEPTIBILITIES DUE TO THE REPAIR OF TYR35 RADICAL BY CYS83 IN Fe-SODB THROUGH INTRAMOLECULAR ELECTRON TRANSFER.
J.Biol.Chem., 289, 2014
|
|
3N3G
 
 | | 4-(3-Trifluoromethylphenyl)-pyrimidine-2-carbonitrile as cathepsin S inhibitors: N3, not N1 is critically important | | Descriptor: | (E)-1-(6-{4-[3-(4-methylpiperazin-1-yl)propoxy]-3-(trifluoromethyl)phenyl}pyridin-2-yl)methanimine, 6-{4-[3-(4-methylpiperazin-1-yl)propoxy]-3-(trifluoromethyl)phenyl}pyridine-2-carbonitrile, Cathepsin S, ... | | Authors: | Fradera, X, Uitdehaag, J.C.M, van Zeeland, M. | | Deposit date: | 2010-05-20 | | Release date: | 2010-07-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | 4-(3-Trifluoromethylphenyl)-pyrimidine-2-carbonitrile as cathepsin S inhibitors: N3, not N1 is critically important.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3MXX
 
 | |
3MX4
 
 | | DNA binding and cleavage by the GIY-YIG endonuclease R.Eco29KI inactive variant E142Q | | Descriptor: | DNA (5'-D(P*CP*GP*GP*GP*AP*GP*GP*CP*CP*CP*GP*CP*GP*GP*GP*CP*CP*GP*CP*CP*GP*C)-3'), DNA (5'-D(P*GP*CP*GP*GP*CP*GP*GP*CP*CP*CP*GP*CP*GP*GP*GP*CP*CP*TP*CP*CP*CP*G)-3'), Eco29kIR | | Authors: | Mak, A.N.S, Lambert, A.R, Stoddard, B.L. | | Deposit date: | 2010-05-06 | | Release date: | 2010-09-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Folding, DNA Recognition, and Function of GIY-YIG Endonucleases: Crystal Structures of R.Eco29kI.
Structure, 18, 2010
|
|
3N0E
 
 | |
6EVV
 
 | | X-ray structure of the complex between human alpha thrombin and NU172, a duplex/quadruplex 26-mer DNA aptamer, in the presence of potassium ions. | | Descriptor: | D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, GLYCEROL, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Troisi, R, Russo Krauss, I, Sica, F. | | Deposit date: | 2017-11-02 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Several structural motifs cooperate in determining the highly effective anti-thrombin activity of NU172 aptamer.
Nucleic Acids Res., 46, 2018
|
|
3NCG
 
 | |
3NZ0
 
 | | Non-phosphorylated TYK2 kinase with CMP6 | | Descriptor: | 2-TERT-BUTYL-9-FLUORO-3,6-DIHYDRO-7H-BENZ[H]-IMIDAZ[4,5-F]ISOQUINOLINE-7-ONE, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2010-07-15 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A new regulatory switch in a JAK protein kinase.
Proteins, 79, 2011
|
|
3O0U
 
 | |
3PDV
 
 | |
3O6G
 
 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | 1,2-ETHANEDIOL, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Maclean, J.K.F, Campbell, R.A, Cumming, I.A, Gillen, K.J, Gillespie, J, Jamieson, C, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Martin, F, Moir, E.M, Morrow, J.A, Pantling, J, Rankovic, Z, Smith, L. | | Deposit date: | 2010-07-29 | | Release date: | 2010-09-15 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel series of positive modulators of the AMPA receptor: structure-based lead optimization.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3OFU
 
 | | Crystal Structure of Cytochrome P450 CYP101C1 | | Descriptor: | (3E)-4-(2,6,6-trimethylcyclohex-1-en-1-yl)but-3-en-2-one, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zhou, W, Ma, M, Bell, S.G, Yang, W, Hao, Y, Rees, N.H, Bartlam, M, Wong, L.-L, Rao, Z. | | Deposit date: | 2010-08-16 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Analysis of CYP101C1 from Novosphingobium aromaticivorans DSM12444.
Chembiochem, 12, 2011
|
|
3ODB
 
 | | Haemophilus influenzae ferric binding protein A -Iron Loaded -open Conformation | | Descriptor: | FE (III) ION, Iron-utilization periplasmic protein | | Authors: | Khambati, H.K, Moraes, T.F, Singh, J, Shouldice, S.R, Yu, R.H, Schryvers, A.B. | | Deposit date: | 2010-08-11 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The role of vicinal tyrosine residues in the function of Haemophilus influenzae ferric binding protein A.
Biochem.J., 2010
|
|
3PIL
 
 | | Crystal structure of Mxr1 from Saccharomyces cerevisiae in reduced form | | Descriptor: | ACETATE ION, Peptide methionine sulfoxide reductase | | Authors: | Ma, X.X, Guo, P.C, Shi, W.W, Luo, M, Tan, X.F, Chen, Y, Zhou, C.Z. | | Deposit date: | 2010-11-07 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural plasticity of the thioredoxin recognition site of yeast methionine S-sulfoxide reductase Mxr1
J.Biol.Chem., 286, 2011
|
|
3PFD
 
 | |
3PH5
 
 | |
3PH6
 
 | |
3NSH
 
 | | BACE-1 in complex with ELN475957 | | Descriptor: | Beta-secretase 1, N-[(1S,2R)-1-(3,5-difluorobenzyl)-3-({1-[4-(2,2-dimethylpropyl)thiophen-2-yl]cyclopropyl}amino)-2-hydroxypropyl]acetamide | | Authors: | Probst, G.D, Bowers, S, Sealy, J.M, Brecht, E, Yao, N. | | Deposit date: | 2010-07-01 | | Release date: | 2010-09-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design and synthesis of hydroxyethylamine (HEA) BACE-1 inhibitors: structure-activity relationship of the aryl region.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3NUD
 
 | | The structure of 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase from mycobacterium tuberculosis complexed with phenylalanine | | Descriptor: | PHENYLALANINE, PHOSPHATE ION, Probable 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase AroG | | Authors: | Parker, E.J, Jameson, G.B, Jiao, W, Webby, C.J, Baker, E.N, Baker, H.M. | | Deposit date: | 2010-07-06 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synergistic allostery, a sophisticated regulatory network for the control of aromatic amino acid biosynthesis in Mycobacterium tuberculosis
J.Biol.Chem., 285, 2010
|
|
3NV6
 
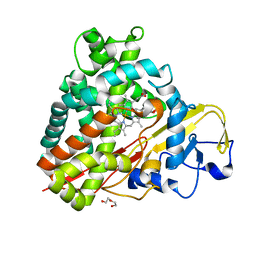 | | Crystal Structure of Camphor-Bound CYP101D2 | | Descriptor: | CAMPHOR, Cytochrome P450, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, W, Bell, S.G, Wang, H, Zhou, W.H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-07-08 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of CYP101D2 unveils a potential path for substrate entry into the active site
Biochem.J., 433, 2011
|
|
3NX2
 
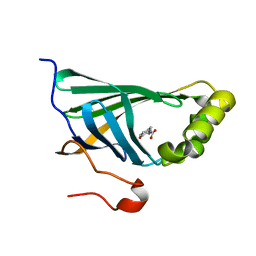 | | Enterobacter sp. Px6-4 Ferulic Acid Decarboxylase in complex with substrate analogues | | Descriptor: | 3-(4-HYDROXY-3-METHOXYPHENYL)-2-PROPENOIC ACID, Ferulic acid decarboxylase | | Authors: | Gu, W, Yang, J.K, Lou, Z.Y, Meng, Z.H, Zhang, K.-Q. | | Deposit date: | 2010-07-12 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Basis of Enzymatic Activity for the Ferulic Acid Decarboxylase (FADase) from Enterobacter sp. Px6-4
Plos One, 6, 2011
|
|
3PX9
 
 | |
6ER6
 
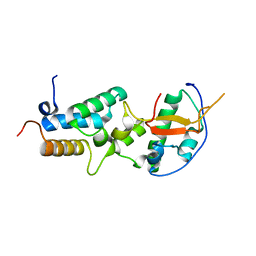 | | Crystal structure of a computationally designed colicin endonuclease and immunity pair colEdes7/Imdes7 | | Descriptor: | Endonuclease colEdes7, immunity Imdes7 | | Authors: | Netzer, R, Listov, D, Dym, O, Albeck, S, Knop, O, Fleishman, S.J. | | Deposit date: | 2017-10-17 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ultrahigh specificity in a network of computationally designed protein-interaction pairs.
Nat Commun, 9, 2018
|
|
