3QI6
 
 | |
3RI6
 
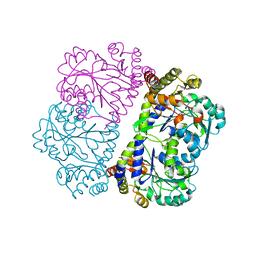 | | A Novel Mechanism of Sulfur Transfer Catalyzed by O-Acetylhomoserine Sulfhydrylase in Methionine Biosynthetic Pathway of Wolinella succinogenes | | Descriptor: | O-ACETYLHOMOSERINE SULFHYDRYLASE | | Authors: | Tran, T.H, Krishnamoorthy, K, Begley, T.P, Ealick, S.E. | | Deposit date: | 2011-04-13 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A novel mechanism of sulfur transfer catalyzed by O-acetylhomoserine sulfhydrylase in the methionine-biosynthetic pathway of Wolinella succinogenes.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
4U2H
 
 | |
8U98
 
 | |
8U99
 
 | |
8DUY
 
 | |
8WKO
 
 | |
8WKR
 
 | |
8ERB
 
 | | Crystal structure of Fub7 in complex with vinylglycine ketimine | | Descriptor: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]but-3-enoic acid, Sulfhydrylase FUB7 | | Authors: | Hai, Y. | | Deposit date: | 2022-10-11 | | Release date: | 2023-10-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Molecular and Structural Basis for C gamma-C Bond Formation by PLP-Dependent Enzyme Fub7.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8EQW
 
 | | Crystal structure of Fub7 | | Descriptor: | Sulfhydrylase FUB7 | | Authors: | Hai, Y. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular and Structural Basis for C gamma-C Bond Formation by PLP-Dependent Enzyme Fub7.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8ERJ
 
 | | Crystal structure of Fub7 in complex with E-2-aminocrotonate | | Descriptor: | (2E)-2-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}but-2-enoic acid, (2S)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]but-3-enoic acid, Sulfhydrylase FUB7 | | Authors: | Hai, Y. | | Deposit date: | 2022-10-12 | | Release date: | 2023-10-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Molecular and Structural Basis for C gamma-C Bond Formation by PLP-Dependent Enzyme Fub7.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
7F1U
 
 | | Crystal structure of Pseudomonas putida methionine gamma-lyase Q349S mutant with L-methionine intermediates | | Descriptor: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]-4-(methylsulfanyl)but-2-enoic acid, L-methionine gamma-lyase, METHIONINE | | Authors: | Okawa, A, Handa, H, Yasuda, E, Murota, M, Kudo, D, Tamura, T, Shiba, T, Inagaki, K. | | Deposit date: | 2021-06-09 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization and application of l-methionine gamma-lyase Q349S mutant enzyme with an enhanced activity toward l-homocysteine.
J.Biosci.Bioeng., 133, 2022
|
|
7F1V
 
 | | Crystal structure of Pseudomonas putida methionine gamma-lyase Q349S mutant with L-homocysteine intermediates | | Descriptor: | (2~{S})-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]-4-sulfanyl-butanoic acid, 2-AMINO-4-MERCAPTO-BUTYRIC ACID, L-methionine gamma-lyase | | Authors: | Okawa, A, Handa, H, Yasuda, E, Murota, M, Kudo, D, Tamura, T, Shiba, T, Inagaki, K. | | Deposit date: | 2021-06-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Characterization and application of l-methionine gamma-lyase Q349S mutant enzyme with an enhanced activity toward l-homocysteine.
J.Biosci.Bioeng., 133, 2022
|
|
7F1P
 
 | | Crystal structure of Pseudomonas putida methionine gamma-lyase Q349S mutant ligand-free form. | | Descriptor: | L-methionine gamma-lyase | | Authors: | Okawa, A, Handa, H, Yasuda, E, Murota, M, Kudo, D, Tamura, T, Shiba, T, Inagaki, K. | | Deposit date: | 2021-06-09 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Characterization and application of l-methionine gamma-lyase Q349S mutant enzyme with an enhanced activity toward l-homocysteine.
J.Biosci.Bioeng., 133, 2022
|
|
4MKJ
 
 | | Crystal structure of L-methionine gamma-lyase from Citrobacter freundii modified by allicine | | Descriptor: | Methionine gamma-lyase, PENTAETHYLENE GLYCOL, SODIUM ION, ... | | Authors: | Revtovich, S.V, Nikulin, A.D, Morozova, E.A, Zakomirdina, L.N, Demidkina, T.V. | | Deposit date: | 2013-09-05 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Alliin is a suicide substrate of Citrobacter freundii methionine gamma-lyase: structural bases of inactivation of the enzyme.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4L0O
 
 | |
1CL2
 
 | |
1CS1
 
 | |
1CL1
 
 | |
7KB0
 
 | | O-acety-L-homoserine aminocarboxypropyltransferase (MetY) from Thermotoga maritima with pyridoxal-5-phosphate (PLP) bound in the internal aldimine state | | Descriptor: | O-acetyl-L-homoserine sulfhydrylase | | Authors: | Brewster, J.L, Pachl, P, Squire, C, Selmer, M, Patrick, W.M. | | Deposit date: | 2020-10-01 | | Release date: | 2021-06-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures and kinetics of Thermotoga maritima MetY reveal new insights into the predominant sulfurylation enzyme of bacterial methionine biosynthesis.
J.Biol.Chem., 296, 2021
|
|
7KB1
 
 | | Complex of O-acety-L-homoserine aminocarboxypropyltransferase (MetY) from Thermotoga maritima and a key reaction intermediate | | Descriptor: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]but-3-enoic acid, MAGNESIUM ION, O-acetyl-L-homoserine sulfhydrylase, ... | | Authors: | Brewster, J.L, Pachl, P, Squire, C, Selmer, M, Patrick, W.M. | | Deposit date: | 2020-10-01 | | Release date: | 2021-06-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures and kinetics of Thermotoga maritima MetY reveal new insights into the predominant sulfurylation enzyme of bacterial methionine biosynthesis.
J.Biol.Chem., 296, 2021
|
|
4OMA
 
 | | The crystal structure of methionine gamma-lyase from Citrobacter freundii in complex with L-cycloserine pyridoxal-5'-phosphate | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL, ... | | Authors: | Revtovich, S.V, Nikulin, A.D, Morozova, E.A, Demidkina, T.V. | | Deposit date: | 2014-01-27 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Pre-steady-state Kinetic and Structural Analysis of Interaction of Methionine gamma-Lyase from Citrobacter freundii with Inhibitors.
J.Biol.Chem., 290, 2015
|
|
4MKK
 
 | | Crystal structure of C115A mutant L-methionine gamma-lyase from Citrobacter freundii modified by allicine | | Descriptor: | CHLORIDE ION, Methionine gamma-lyase, POTASSIUM ION, ... | | Authors: | Revtovich, S.V, Nikulin, A.D, Morozova, E.A, Zakomirdina, L.N, Demidkina, T.V. | | Deposit date: | 2013-09-05 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Alliin is a suicide substrate of Citrobacter freundii methionine gamma-lyase: structural bases of inactivation of the enzyme.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4OC9
 
 | | 2.35 Angstrom resolution crystal structure of putative O-acetylhomoserine (thiol)-lyase (metY) from Campylobacter jejuni subsp. jejuni NCTC 11168 with N'-Pyridoxyl-Lysine-5'-Monophosphate at position 205 | | Descriptor: | GLYCEROL, IMIDAZOLE, PHOSPHATE ION, ... | | Authors: | Halavaty, A.S, Brunzelle, J.S, Wawrzak, Z, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-08 | | Release date: | 2014-03-12 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2.35 Angstrom resolution crystal structure of putative O-acetylhomoserine (thiol)-lyase (metY) from Campylobacter jejuni subsp. jejuni NCTC 11168 with N'-Pyridoxyl-Lysine-5'-Monophosphate at position 205
To be Published
|
|
1E5F
 
 | | METHIONINE GAMMA-LYASE (MGL) FROM TRICHOMONAS VAGINALIS | | Descriptor: | GLYCEROL, METHIONINE GAMMA-LYASE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Goodall, G, Mottram, J.C, Coombs, G.H, Lapthorn, A.J. | | Deposit date: | 2000-07-25 | | Release date: | 2001-07-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | The Structure and Proposed Catalytic Mechanism of Methionine Gamma-Lyase
To be Published
|
|
