3IF8
 
 | |
3IGP
 
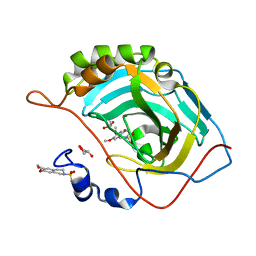 | | Structure of inhibitor binding to Carbonic Anhydrase II | | Descriptor: | 6,7-dimethoxy-3,4-dihydroisoquinoline-2(1H)-sulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Gitto, R, Agnello, S, Brynda, J, Mader, P, Supuran, C.T, Chimirri, A. | | Deposit date: | 2009-07-28 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of 3,4-Dihydroisoquinoline-2(1H)-sulfonamides as Potent Carbonic Anhydrase Inhibitors: Synthesis, Biological Evaluation, and Enzyme-Ligand X-ray Studies.
J.Med.Chem., 2010
|
|
3II5
 
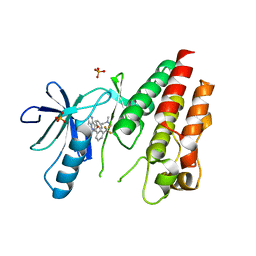 | | The Complex of wild-type B-RAF with Pyrazolo pyrimidine inhibitor | | Descriptor: | B-Raf proto-oncogene serine/threonine-protein kinase, N-[3-(3-{4-[(dimethylamino)methyl]phenyl}pyrazolo[1,5-a]pyrimidin-7-yl)phenyl]-3-(trifluoromethyl)benzamide, PHOSPHATE ION | | Authors: | Xu, W, Breger, D, Torres, N, Dutia, M, Powell, D, Ciszewski, G. | | Deposit date: | 2009-07-31 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Non-hinge-binding pyrazolo[1,5-a]pyrimidines as potent B-Raf kinase inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3IW6
 
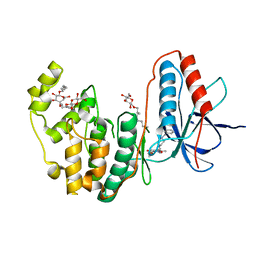 | | Human p38 MAP Kinase in Complex with a Benzylpiperazin-Pyrrol | | Descriptor: | Mitogen-activated protein kinase 14, ethyl 4-[(4-benzylpiperazin-1-yl)carbonyl]-1-ethyl-3,5-dimethyl-1H-pyrrole-2-carboxylate, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Rauh, D. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-Throughput Screening To Identify Inhibitors Which Stabilize Inactive Kinase Conformations in p38alpha
J.Am.Chem.Soc., 131, 2009
|
|
3I7O
 
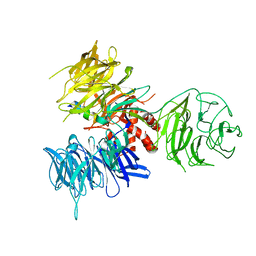 | | Crystal Structure of DDB1 in Complex with the H-Box Motif of IQWD1 | | Descriptor: | DNA damage-binding protein 1, IQ motif and WD repeat-containing protein 1 | | Authors: | Li, T, Robert, E.I, Breugel, P.C.V, Strubin, M, Zheng, N. | | Deposit date: | 2009-07-08 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3IMW
 
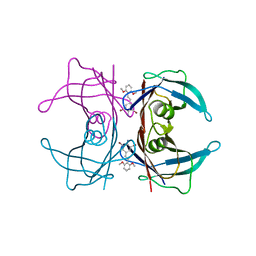 | | Transthyretin in complex with (E)-2,6-dibromo-4-(2,6-dimethoxystyryl)aniline | | Descriptor: | 2,6-dibromo-4-[(E)-2-(2,6-dimethoxyphenyl)ethenyl]aniline, Transthyretin | | Authors: | Connelly, S, Wilson, I.A. | | Deposit date: | 2009-08-11 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | A substructure combination strategy to create potent and selective transthyretin kinetic stabilizers that prevent amyloidogenesis and cytotoxicity.
J.Am.Chem.Soc., 132, 2010
|
|
3I97
 
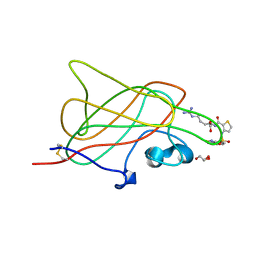 | | B1 domain of human Neuropilin-1 bound with small molecule EG00229 | | Descriptor: | (S)-2-(3-(benzo[c][1,2,5]thiadiazole-4-sulfonamido)thiophene-2-carboxamido)-5-guanidinopentanoic acid, GLYCEROL, Neuropilin-1 | | Authors: | Allerston, C.K, Djordjevic, S. | | Deposit date: | 2009-07-10 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Small molecule inhibitors of the neuropilin-1 vascular endothelial growth factor A (VEGF-A) interaction.
J.Med.Chem., 53, 2010
|
|
3IO7
 
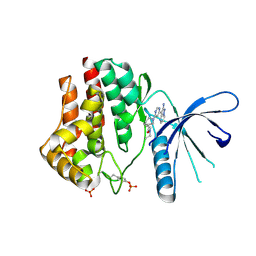 | | 2-Aminopyrazolo[1,5-a]pyrimidines as potent and selective inhibitors of JAK2 | | Descriptor: | (3S)-1-[6-(2-aminopyrazolo[1,5-a]pyrimidin-3-yl)pyrimidin-4-yl]-N,N-diethylpiperidine-3-carboxamide, Tyrosine-protein kinase JAK2 | | Authors: | Zuccola, H.J, Ledeboer, M.W, Pierce, A.C. | | Deposit date: | 2009-08-13 | | Release date: | 2009-11-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2-Aminopyrazolo[1,5-a]pyrimidines as potent and selective inhibitors of JAK2.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3IOJ
 
 | | Crystal structure of the Fucosylgalactoside alpha N-acetylgalactosaminyltransferase (GTA, cisAB mutant L266G, G268A) in complex with UDP | | Descriptor: | GLYCEROL, Histo-blood group ABO system transferase, MANGANESE (II) ION, ... | | Authors: | Pesnot, T, Jorgensen, R, Palcic, M.M, Wagner, G.K. | | Deposit date: | 2009-08-14 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Structural and mechanistic basis for a new mode of glycosyltransferase inhibition.
Nat.Chem.Biol., 6, 2010
|
|
3IFQ
 
 | | Interction of plakoglobin and beta-catenin with desmosomal cadherins | | Descriptor: | E-cadherin, SULFATE ION, plakoglobin | | Authors: | Choi, H.-J, Gross, J.C, Pokutta, S, Weis, W.I. | | Deposit date: | 2009-07-24 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Interactions of plakoglobin and beta-catenin with desmosomal cadherins: basis of selective exclusion of alpha- and beta-catenin from desmosomes.
J.Biol.Chem., 284, 2009
|
|
3IO4
 
 | | Huntingtin amino-terminal region with 17 Gln residues - Crystal C90 | | Descriptor: | CALCIUM ION, Maltose-binding periplasmic protein,Huntingtin fusion protein, ZINC ION | | Authors: | Kim, M.W, Chelliah, Y, Kim, S.W, Otwinowski, Z, Bezprozvanny, I. | | Deposit date: | 2009-08-13 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.63 Å) | | Cite: | Secondary structure of Huntingtin amino-terminal region.
Structure, 17, 2009
|
|
3IMS
 
 | | Transthyretin in complex with 2,6-dibromo-4-(2,6-dichlorophenethyl)phenol | | Descriptor: | 2,6-dibromo-4-[2-(2,6-dichlorophenyl)ethyl]phenol, Transthyretin | | Authors: | Connelly, S, Wilson, I.A. | | Deposit date: | 2009-08-11 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A substructure combination strategy to create potent and selective transthyretin kinetic stabilizers that prevent amyloidogenesis and cytotoxicity.
J.Am.Chem.Soc., 132, 2010
|
|
3IN5
 
 | | Structure of human DNA polymerase kappa inserting dATP opposite an 8-oxoG DNA lesion | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*C*CP*TP*AP*(8OG)P*GP*AP*GP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3'), DNA (5'-D(*GP*G*GP*GP*GP*AP*AP*GP*GP*AP*CP*TP*(DOC))-3'), ... | | Authors: | Silverstein, T.D, Vasquez-Del Carpio, R, Aggarwal, A.K. | | Deposit date: | 2009-08-11 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of human DNA polymerase kappa inserting dATP opposite an 8-oxoG DNA lesion
PLOS ONE, 4, 2009
|
|
3IOI
 
 | | Crystal structure of the Fucosylgalactoside alpha N-acetylgalactosaminyltransferase (GTA, cisAB mutant L266G, G268A) in complex with a novel UDP-Gal derived inhibitor (1GW) | | Descriptor: | 5-(2-FORMYLTHIEN-5-YL)-URIDINE-5'-DIPHOSPHATE-ALPHA-D-GALACTOSE, Histo-blood group ABO system transferase, MANGANESE (II) ION, ... | | Authors: | Pesnot, T, Jorgensen, R, Palcic, M.M, Wagner, G.K. | | Deposit date: | 2009-08-14 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and mechanistic basis for a new mode of glycosyltransferase inhibition.
Nat.Chem.Biol., 6, 2010
|
|
3IRT
 
 | |
3ISC
 
 | | Binary complex of human DNA polymerase beta with an abasic site (THF) in the gapped DNA | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*(3DR)P*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Beard, W.A, Shock, D.D, Batra, V.K, Pedersen, L.C, Wilson, S.H. | | Deposit date: | 2009-08-25 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | DNA polymerase beta substrate specificity: side chain modulation of the "A-rule".
J.Biol.Chem., 284, 2009
|
|
3IRQ
 
 | | Crystal structure of a Z-Z junction | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*CP*GP*CP*GP*AP*CP*GP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*TP*CP*GP*CP*GP*CP*GP*TP*CP*GP*CP*GP*CP*G)-3'), Double-stranded RNA-specific adenosine deaminase | | Authors: | Athanasiadis, A, de Rosa, M. | | Deposit date: | 2009-08-24 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a junction between two Z-DNA helices.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3ISW
 
 | | Crystal structure of filamin-A immunoglobulin-like repeat 21 bound to an N-terminal peptide of CFTR | | Descriptor: | Cystic fibrosis transmembrane conductance regulator, Filamin-A | | Authors: | Xu, Z, Page, R, Qin, J, Ithychanda, S.S, Liu, J.M, Misra, S. | | Deposit date: | 2009-08-27 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Biochemical basis of the interaction between cystic fibrosis transmembrane conductance regulator and immunoglobulin-like repeats of filamin.
J.Biol.Chem., 285, 2010
|
|
3ISD
 
 | | Ternary complex of human DNA polymerase beta with an abasic site (THF): DAPCPP mismatch | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxy-5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]adenosine, 5'-D(*CP*CP*GP*AP*CP*(3DR)P*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', ... | | Authors: | Beard, W.A, Shock, D.D, Batra, V.K, Pedersen, L.C, Wilson, S.H. | | Deposit date: | 2009-08-25 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | DNA polymerase beta substrate specificity: side chain modulation of the "A-rule".
J.Biol.Chem., 284, 2009
|
|
3I7N
 
 | | Crystal Structure of DDB1 in Complex with the H-Box Motif of WDTC1 | | Descriptor: | DNA damage-binding protein 1, WD and tetratricopeptide repeats protein 1 | | Authors: | Li, T, Robert, E.I, Breugel, P.C.V, Strubin, M, Zheng, N. | | Deposit date: | 2009-07-08 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3IPS
 
 | | X-ray structure of benzisoxazole synthetic agonist bound to the LXR-alpha | | Descriptor: | Nuclear receptor coactivator 1, Oxysterols receptor LXR-alpha, SULFATE ION, ... | | Authors: | Fradera, X, Vu, D, Nimz, O, Skene, R, Hosfield, D, Wijnands, R, Cooke, A.J, Haunso, A, King, A, Bennet, D.J, McGuire, R, Uitdehaag, J.C.M. | | Deposit date: | 2009-08-18 | | Release date: | 2010-06-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | X-ray structures of the LXRalpha LBD in its homodimeric form and implications for heterodimer signaling.
J.Mol.Biol., 399, 2010
|
|
3IQS
 
 | | Crystal structure of the anti-viral APOBEC3G catalytic domain | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, ZINC ION | | Authors: | Holden, L.G, Prochnow, C, Chang, Y.P, Bransteitter, R, Chelico, L, Sen, U, Stevens, R.C, Goodman, R.F, Chen, X.S. | | Deposit date: | 2009-08-20 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications.
Nature, 456, 2008
|
|
3I7H
 
 | | Crystal Structure of DDB1 in Complex with the H-Box Motif of HBX | | Descriptor: | DNA damage-binding protein 1, X protein | | Authors: | Li, T, Robert, E.I, Breugel, P.C.V, Strubin, M, Zheng, N. | | Deposit date: | 2009-07-08 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3I80
 
 | | Protein Tyrosine Phosphatase 1B - Transition state analog for the second catalytic step | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Tyrosine-protein phosphatase non-receptor type 1, ... | | Authors: | Brandao, T.A.S, Johnson, S.J, Hengge, A.C. | | Deposit date: | 2009-07-09 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Insights into the reaction of protein-tyrosine phosphatase 1B: crystal structures for transition state analogs of both catalytic steps.
J.Biol.Chem., 285, 2010
|
|
3I9M
 
 | | Crystal structure of human CD38 complexed with an analog ara-2'F-ADPR | | Descriptor: | ADP-ribosyl cyclase 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3R,4R)-4-fluoro-3-hydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate | | Authors: | Liu, Q, Graeff, R, Kriksunov, I.A, Jiang, H, Zhang, B, Oppenheimer, N, Lin, H, Potter, B.V.L, Lee, H.C, Hao, Q. | | Deposit date: | 2009-07-12 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for enzymatic evolution from a dedicated ADP-ribosyl cyclase to a multifunctional NAD hydrolase
J.Biol.Chem., 284, 2009
|
|
