3BEA
 
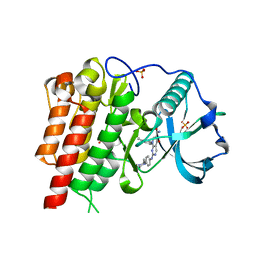 | | cFMS tyrosine kinase (tie2 KID) in complex with a pyrimidinopyridone inhibitor | | Descriptor: | 8-(2,3-dihydro-1H-inden-5-yl)-2-({4-[(3R,5S)-3,5-dimethylpiperazin-1-yl]phenyl}amino)-5-oxo-5,8-dihydropyrido[2,3-d]pyrimidine-6-carboxamide, Macrophage colony-stimulating factor 1 receptor, SULFATE ION | | Authors: | Schubert, C. | | Deposit date: | 2007-11-16 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Design and synthesis of a pyrido[2,3-d]pyrimidin-5-one class of anti-inflammatory FMS inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3BEJ
 
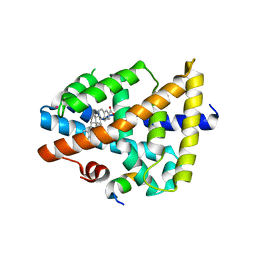 | | Structure of human FXR in complex with MFA-1 and co-activator peptide | | Descriptor: | (8alpha,10alpha,13alpha,17beta)-17-[(4-hydroxyphenyl)carbonyl]androsta-3,5-diene-3-carboxylic acid, Bile acid receptor, Nuclear receptor coactivator 1, ... | | Authors: | Soisson, S.M, Parthasarathy, G, Becker, J.W. | | Deposit date: | 2007-11-19 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of a potent synthetic FXR agonist with an unexpected mode of binding and activation.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3BEW
 
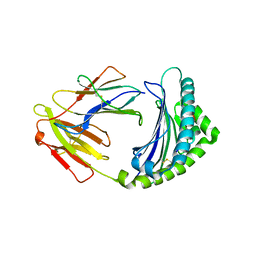 | | 10mer Crystal Structure of chicken MHC class I haplotype B21 | | Descriptor: | 10-mer from Tubulin beta-6 chain, Beta-2-microglobulin, Major histocompatibility complex class I glycoprotein haplotype B21 | | Authors: | Koch, M, Camp, S, Collen, T, Avila, D, Salomonsen, J, Wallny, H.J, van Hateren, A, Hunt, L, Jacob, J.P, Johnston, F, Marston, D.A, Shaw, I, Dunbar, P.R, Cerundolo, V, Jones, E.Y, Kaufman, J. | | Deposit date: | 2007-11-20 | | Release date: | 2008-01-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of an MHC class I molecule from b21 chickens illustrate promiscuous Peptide binding
Immunity, 27, 2007
|
|
3BIA
 
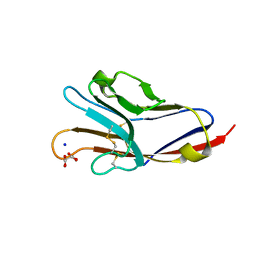 | | Tim-4 in complex with sodium potassium tartrate | | Descriptor: | L(+)-TARTARIC ACID, SODIUM ION, T-cell immunoglobulin and mucin domain-containing protein 4 | | Authors: | Santiago, C, Ballesteros, A, Kaplan, G.G, Freeman, G.J, Casasnovas, J.M. | | Deposit date: | 2007-11-30 | | Release date: | 2008-01-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of T Cell Immunoglobulin Mucin Protein 4 Show a Metal-Ion-Dependent Ligand Binding Site where Phosphatidylserine Binds.
Immunity, 27, 2007
|
|
2J2S
 
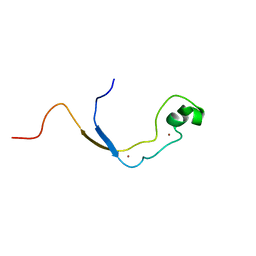 | | Solution structure of the nonmethyl-CpG-binding CXXC domain of the leukaemia-associated MLL histone methyltransferase | | Descriptor: | ZINC FINGER PROTEIN HRX, ZINC ION | | Authors: | Allen, M.D, Grummitt, C.G, Hilcenko, C, Young-Min, S, Tonkin, L.M, Johnson, C.M, Bycroft, M, Warren, A.J. | | Deposit date: | 2006-08-17 | | Release date: | 2006-08-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Nonmethyl-Cpg-Binding Cxxc Domain of the Leukaemia-Associated Mll Histone Methyltransferase
Embo J., 25, 2006
|
|
3B2T
 
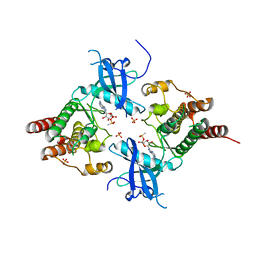 | | Structure of phosphotransferase | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(methyl)phosphoryl]oxy}phosphoryl]adenosine, Fibroblast growth factor receptor 2, PHOSPHATE ION | | Authors: | Lew, E.D, Bae, J.H, Rohmann, E, Wollnik, B, Schlessinger, J. | | Deposit date: | 2007-10-19 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for reduced FGFR2 activity in LADD syndrome: Implications for FGFR autoinhibition and activation.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3B3S
 
 | | Crystal structure of the M180A mutant of the aminopeptidase from Vibrio proteolyticus in complex with leucine | | Descriptor: | Bacterial leucyl aminopeptidase, LEUCINE, SODIUM ION, ... | | Authors: | Ataie, N.J, Hoang, Q.Q, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-10-22 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Zinc coordination geometry and ligand binding affinity: the structural and kinetic analysis of the second-shell serine 228 residue and the methionine 180 residue of the aminopeptidase from Vibrio proteolyticus.
Biochemistry, 47, 2008
|
|
3B3C
 
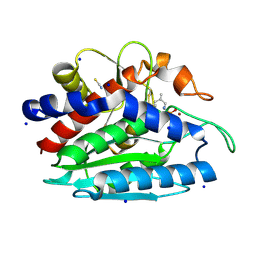 | | Crystal structure of the M180A mutant of the aminopeptidase from Vibrio proteolyticus in complex with leucine phosphonic acid | | Descriptor: | Bacterial leucyl aminopeptidase, LEUCINE PHOSPHONIC ACID, POTASSIUM ION, ... | | Authors: | Ataie, N.J, Hoang, Q.Q, Petsko, G.A, Ringe, D. | | Deposit date: | 2007-10-19 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Zinc coordination geometry and ligand binding affinity: the structural and kinetic analysis of the second-shell serine 228 residue and the methionine 180 residue of the aminopeptidase from Vibrio proteolyticus.
Biochemistry, 47, 2008
|
|
3B67
 
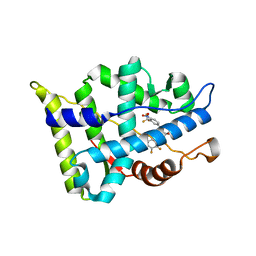 | |
3B65
 
 | |
3B75
 
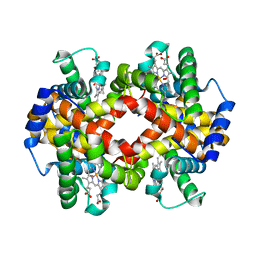 | | Crystal Structure of Glycated Human Haemoglobin | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, OXYGEN MOLECULE, ... | | Authors: | Saraswathi, N.T, Syakhovich, V.E, Bokut, S.B, Moras, D, Ruff, M. | | Deposit date: | 2007-10-30 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The effect of hemoglobin glycosylation on diabete linked oxidative stress
To be Published
|
|
2IVT
 
 | |
3B7B
 
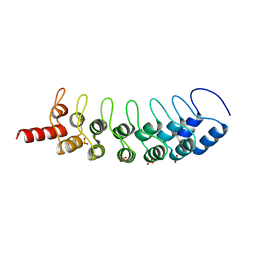 | | EuHMT1 (Glp) Ankyrin Repeat Domain (Structure 1) | | Descriptor: | Euchromatic histone-lysine N-methyltransferase 1, SULFATE ION | | Authors: | Collins, R.E, Horton, J.R, Cheng, X. | | Deposit date: | 2007-10-30 | | Release date: | 2008-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The ankyrin repeats of G9a and GLP histone methyltransferases are mono- and dimethyllysine binding modules
Nat.Struct.Mol.Biol., 15, 2008
|
|
3B92
 
 | |
2J2P
 
 | | L-ficolin complexed to N-acetyl-cystein (150mM) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, FICOLIN-2, ... | | Authors: | Garlatti, V, Gaboriaud, C. | | Deposit date: | 2006-08-17 | | Release date: | 2007-01-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights Into the Innate Immune Recognition Specificities of L- and H-Ficolins.
Embo J., 26, 2007
|
|
2ITU
 
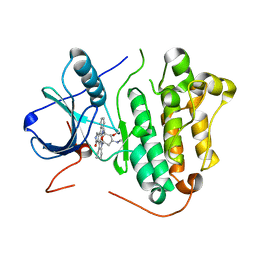 | | Crystal structure of EGFR kinase domain L858R mutation in complex with AFN941 | | Descriptor: | 1,2,3,4-Tetrahydrogen Staurosporine, EPIDERMAL GROWTH FACTOR RECEPTOR | | Authors: | Yun, C.-H, Boggon, T.J, Li, Y, Woo, S, Greulich, H, Meyerson, M, Eck, M.J. | | Deposit date: | 2006-05-25 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Lung Cancer-Derived Egfr Mutants and Inhibitor Complexes: Mechanism of Activation and Insights Into Differential Inhibitor Sensitivity
Cancer Cell, 11, 2007
|
|
3BAQ
 
 | |
3BA5
 
 | |
3BAD
 
 | |
2J5E
 
 | |
3BEU
 
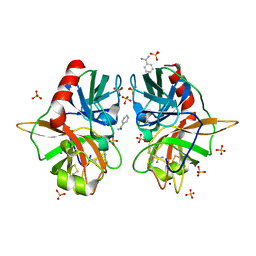 | | Na+-Dependent Allostery Mediates Coagulation Factor Protease Active Site Selectivity | | Descriptor: | BENZAMIDINE, CALCIUM ION, SODIUM ION, ... | | Authors: | Page, M.J, Carrell, C.J, Di Cera, E. | | Deposit date: | 2007-11-20 | | Release date: | 2008-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Engineering protein allostery: 1.05 A resolution structure and enzymatic properties of a Na+-activated trypsin.
J.Mol.Biol., 378, 2008
|
|
2VLE
 
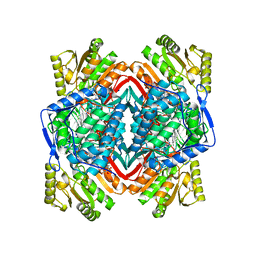 | | The structure of daidzin, a naturally occurring anti alcohol- addiction agent, in complex with human mitochondrial aldehyde dehydrogenase | | Descriptor: | ALDEHYDE DEHYDROGENASE, MITOCHONDRIAL, DAIDZIN | | Authors: | Lowe, E.D, Gao, G.Y, Johnson, L.N, Keung, W.M. | | Deposit date: | 2008-01-13 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Daidzin, a Naturally Occurring Anti-Alcohol-Addiction Agent, in Complex with Human Mitochondrial Aldehyde Dehydrogenase.
J.Med.Chem., 51, 2008
|
|
2V14
 
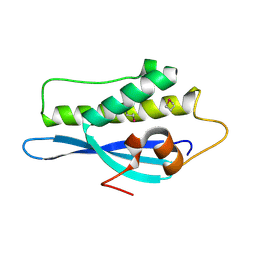 | | Kinesin 16B Phox-homology domain (KIF16B) | | Descriptor: | KINESIN-LIKE MOTOR PROTEIN C20ORF23 | | Authors: | Wilson, M.I, Williams, R.L, Cho, W, Hong, W, Blatner, N.R. | | Deposit date: | 2007-05-21 | | Release date: | 2007-07-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structural Basis of Novel Endosome Anchoring Activity of Kif16B Kinesin.
Embo J., 26, 2007
|
|
3BEL
 
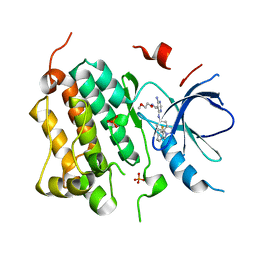 | | X-ray structure of EGFR in complex with oxime inhibitor | | Descriptor: | 4-amino-6-{[1-(3-fluorobenzyl)-1H-indazol-5-yl]amino}pyrimidine-5-carbaldehyde O-(2-methoxyethyl)oxime, Epidermal growth factor receptor, PHOSPHATE ION | | Authors: | Abad, M.C, Xu, G, Neeper, M.P, Struble, G.T, Gaul, M.D, Connolly, P.J. | | Deposit date: | 2007-11-19 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of novel 4-amino-6-arylaminopyrimidine-5-carbaldehyde oximes as dual inhibitors of EGFR and ErbB-2 protein tyrosine kinases.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3GE2
 
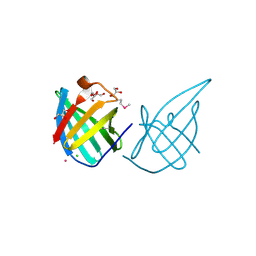 | | Crystal structure of putative lipoprotein SP_0198 from Streptococcus pneumoniae | | Descriptor: | CHLORIDE ION, GLYCEROL, Lipoprotein, ... | | Authors: | Kim, Y, Zhang, R, Joachimiak, G, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-24 | | Release date: | 2009-03-17 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.203 Å) | | Cite: | Crystal Structure of Putative Lipoprotein SP_0198 from Streptococcus pneumoniae
To be Published
|
|
