2CMA
 
 | | Structural Basis for Inhibition of Protein Tyrosine Phosphatase 1B by Isothiazolidinone Heterocyclic Phosphonate Mimetics | | Descriptor: | N-BENZOYL-L-PHENYLALANYL-4-[(5S)-1,1-DIOXIDO-3-OXOISOTHIAZOLIDIN-5-YL]-L-PHENYLALANINAMIDE, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Ala, P.J, Gonneville, L, Hillman, M.C, Becker-Pasha, M, Wei, M, Reid, B.G, Klabe, R, Yue, E.W, Wayland, B, Douty, B, Combs, A.P, Polam, P, Wasserman, Z, Bower, M, Burn, T.C, Hollis, G.F, Wynn, R. | | Deposit date: | 2006-05-04 | | Release date: | 2006-08-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Inhibition of Protein-Tyrosine Phosphatase 1B by Isothiazolidinone Heterocyclic Phosphonate Mimetics.
J.Biol.Chem., 281, 2006
|
|
4GLV
 
 | | OBody AM3L09 bound to hen egg-white lysozyme | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Lysozyme C, ... | | Authors: | Steemson, J.D. | | Deposit date: | 2012-08-15 | | Release date: | 2013-08-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.574 Å) | | Cite: | Tracking Molecular Recognition at the Atomic Level with a New Protein Scaffold Based on the OB-Fold.
Plos One, 9, 2014
|
|
4JVY
 
 | | Structure of the STAR (signal transduction and activation of RNA) domain of GLD-1 bound to RNA | | Descriptor: | Female germline-specific tumor suppressor gld-1, RNA (5'-R(P*CP*UP*AP*AP*CP*AP*A)-3') | | Authors: | Teplova, M, Hafner, M, Teplov, D, Essig, K, Tuschl, T, Patel, D.J. | | Deposit date: | 2013-03-26 | | Release date: | 2013-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.853 Å) | | Cite: | Structure-function studies of STAR family Quaking proteins bound to their in vivo RNA target sites.
Genes Dev., 27, 2013
|
|
2M4L
 
 | |
6LSR
 
 | | Cryo-EM structure of a pre-60S ribosomal subunit - state B | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Liang, X, Zuo, M, Zhang, Y, Li, N, Ma, C, Dong, M, Gao, N. | | Deposit date: | 2020-01-20 | | Release date: | 2020-08-26 | | Last modified: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural snapshots of human pre-60S ribosomal particles before and after nuclear export.
Nat Commun, 11, 2020
|
|
1K92
 
 | |
2C9S
 
 | | 1.24 Angstroms resolution structure of Zn-Zn Human Superoxide dismutase | | Descriptor: | ACETATE ION, SULFATE ION, SUPEROXIDE DISMUTASE [CU-ZN], ... | | Authors: | Strange, R.W, Antonyuk, S.V, Hough, M.A, Doucette, P.A, Valentine S, J.S, Hasnain, S. | | Deposit date: | 2005-12-14 | | Release date: | 2005-12-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Variable Metallation of Human Superoxide Dismutase: Atomic Resolution Crystal Structures of Cu-Zn, Zn-Zn and as-Isolated Wild-Type Enzymes.
J.Mol.Biol., 356, 2006
|
|
2QMW
 
 | | The crystal structure of the prephenate dehydratase (PDT) from Staphylococcus aureus subsp. aureus Mu50 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tan, K, Zhang, R, Li, H, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-17 | | Release date: | 2007-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of open (R) and close (T) states of prephenate dehydratase (PDT) - implication of allosteric regulation by L-phenylalanine.
J.Struct.Biol., 162, 2008
|
|
7FU0
 
 | | Crystal Structure of human cyclic GMP-AMP synthase in complex with 2-[(4-phenylphenyl)methylamino]-5-(trifluoromethyl)-4H-[1,2,4]triazolo[1,5-a]pyrimidin-7-one | | Descriptor: | (8S)-2-{[([1,1'-biphenyl]-4-yl)methyl]amino}-5-(trifluoromethyl)[1,2,4]triazolo[1,5-a]pyrimidin-7(4H)-one, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Leibrock, L, Benz, J, Groebke-Zbinden, K, Hunziker, D, Rudolph, M.G. | | Deposit date: | 2023-02-08 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Crystal Structure of a human cyclic GMP-AMP synthase complex
To be published
|
|
6M07
 
 | | Crystal structure of Lp-PLA2 in complex with a novel covalent inhibitor | | Descriptor: | (2S)-2-[(E)-3-[2-(diethylamino)ethyl-[[4-[4-(trifluoromethyl)-2-[2,2,2-tris(fluoranyl)ethoxy]phenyl]phenyl]methyl]amino]-1-oxidanyl-3-oxidanylidene-prop-1-enyl]pyrrolidine-1-carboxylic acid, Platelet-activating factor acetylhydrolase | | Authors: | Hu, H.C, Xu, Y.C. | | Deposit date: | 2020-02-20 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Identification of Highly Selective Lipoprotein-Associated Phospholipase A2 (Lp-PLA2) Inhibitors by a Covalent Fragment-Based Approach.
J.Med.Chem., 63, 2020
|
|
3S0K
 
 | | Crystal Structure of Human Glycolipid Transfer Protein complexed with glucosylceramide containing oleoyl acyl chain (18:1) | | Descriptor: | (9Z)-N-[(2S,3R,4E)-1-(beta-D-glucopyranosyloxy)-3-hydroxyoctadec-4-en-2-yl]octadec-9-enamide, Glycolipid transfer protein, NICKEL (II) ION | | Authors: | Cabo-Bilbao, A, Samygina, V, Popov, A.N, Ochoa-Lizarralde, B, Goni-de-Cerio, F, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2011-05-13 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein.
Structure, 19, 2011
|
|
4BY5
 
 | |
1DYB
 
 | |
4KGH
 
 | |
2QHR
 
 | | Crystal structure of the 13F6-1-2 Fab fragment bound to its Ebola virus glycoprotein peptide epitope. | | Descriptor: | 13F6-1-2 Fab fragment V lambda x light chain, 13F6-1-2 Fab fragment heavy chain, Envelope glycoprotein peptide | | Authors: | Lee, J.E, Kuehne, A, Abelson, D.M, Fusco, M.L, Hart, M.K, Saphire, E.O. | | Deposit date: | 2007-07-02 | | Release date: | 2008-01-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complex of a protective antibody with its Ebola virus GP peptide epitope: unusual features of a V lambda x light chain.
J.Mol.Biol., 375, 2008
|
|
6MEP
 
 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC3437 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Tyrosine-protein kinase Mer, ... | | Authors: | Da, C, Zhang, D, Stashko, M.A, Cheng, A, Hunter, D, Norris-Drouin, J, Graves, L, Machius, M, Miley, M.J, DeRyckere, D, Earp, H.S, Graham, D.K, Frye, S.V, Wang, X, Kireev, D. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.893 Å) | | Cite: | Data-Driven Construction of Antitumor Agents with Controlled Polypharmacology.
J.Am.Chem.Soc., 141, 2019
|
|
1SNB
 
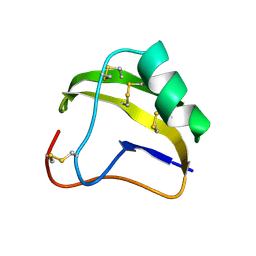 | | STRUCTURE OF SCORPION NEUROTOXIN BMK M8 | | Descriptor: | NEUROTOXIN BMK M8 | | Authors: | Wang, D.C, Zeng, Z.H, Li, H.M. | | Deposit date: | 1997-03-12 | | Release date: | 1997-05-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an acidic neurotoxin from scorpion Buthus martensii Karsch at 1.85 A resolution.
J.Mol.Biol., 261, 1996
|
|
4GMX
 
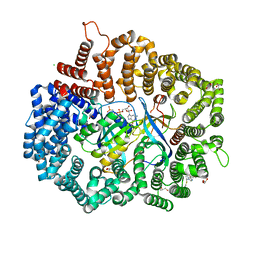 | |
1P0M
 
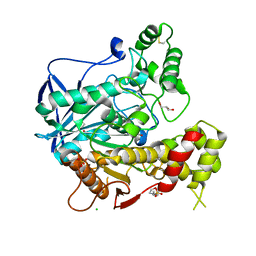 | | Crystal structure of human butyryl cholinesterase in complex with a choline molecule | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nicolet, Y, Lockridge, O, Masson, P, Fontecilla-Camps, J.C, Nachon, F. | | Deposit date: | 2003-04-10 | | Release date: | 2003-08-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of human butyrylcholinesterase and of its complexes with substrate and products.
J.Biol.Chem., 278, 2003
|
|
2DAJ
 
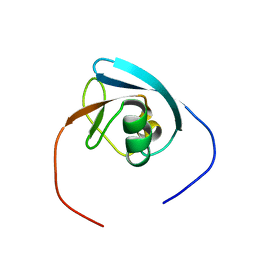 | | Solution Structure of the Novel Identified Ubiquitin-like Domain in the Human COBL-like 1 Protein | | Descriptor: | KIAA0977 protein | | Authors: | Zhao, C, Kigawa, T, Saito, K, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-14 | | Release date: | 2006-06-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Novel Identified Ubiquitin-like Domain in the Human COBL-like 1 Protein
To be Published
|
|
1OVZ
 
 | | Crystal structure of human FcaRI | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Herr, A.B, Ballister, E.R, Bjorkman, P.J. | | Deposit date: | 2003-03-27 | | Release date: | 2003-05-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Insights into IgA-mediated immune responses from the crystal structures of human Fc-alpha-RI and its complex with IgA1-Fc
Nature, 423, 2003
|
|
6M06
 
 | | Crystal structure of Lp-PLA2 in complex with a novel covalent inhibitor | | Descriptor: | (2S)-2-[(Z)-1,3-bis(oxidanyl)-3-oxidanylidene-prop-1-enyl]pyrrolidine-1-carboxylic acid, Platelet-activating factor acetylhydrolase | | Authors: | Hu, H.C, Xu, Y.C. | | Deposit date: | 2020-02-20 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of Highly Selective Lipoprotein-Associated Phospholipase A2 (Lp-PLA2) Inhibitors by a Covalent Fragment-Based Approach.
J.Med.Chem., 63, 2020
|
|
3SJQ
 
 | | Crystal structure of a small conductance potassium channel splice variant complexed with calcium-calmodulin | | Descriptor: | 1-phenylurea, CALCIUM ION, Calmodulin, ... | | Authors: | Zhang, M, Pascal, J.M, Zhang, J.-F. | | Deposit date: | 2011-06-21 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for calmodulin as a dynamic calcium sensor.
Structure, 20, 2012
|
|
2D1Y
 
 | |
2QWP
 
 | | Crystal structure of disulfide-bond-crosslinked complex of bovine hsc70 (1-394aa)R171C and bovine Auxilin (810-910aa)D876C in the ADP*Pi form #2 | | Descriptor: | ACETIC ACID, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Jiang, J, Maes, E.G, Wang, L, Taylor, A.B, Hinck, A.P, Lafer, E.M, Sousa, R. | | Deposit date: | 2007-08-10 | | Release date: | 2007-12-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of J cochaperone binding and regulation of Hsp70.
Mol.Cell, 28, 2007
|
|
