1A7T
 
 | | METALLO-BETA-LACTAMASE WITH MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, METALLO-BETA-LACTAMASE, SODIUM ION, ... | | Authors: | Fitzgerald, P.M.D, Wu, J.K, Toney, J.H. | | Deposit date: | 1998-03-17 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Unanticipated inhibition of the metallo-beta-lactamase from Bacteroides fragilis by 4-morpholineethanesulfonic acid (MES): a crystallographic study at 1.85-A resolution.
Biochemistry, 37, 1998
|
|
6K05
 
 | | Crystal structure of BRD2(BD1)with ligand BY27 bound | | Descriptor: | (6~{R})-~{N}-(4-chlorophenyl)-1-methyl-8-(1-methylpyrazol-4-yl)-5,6-dihydro-4~{H}-[1,2,4]triazolo[4,3-a][1]benzazepin-6-amine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bromodomain-containing protein 2 | | Authors: | Lu, T, Lu, W, Chen, D, Zhao, Y, Luo, C. | | Deposit date: | 2019-05-05 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Discovery, structural insight, and bioactivities of BY27 as a selective inhibitor of the second bromodomains of BET proteins.
Eur.J.Med.Chem., 182, 2019
|
|
2BRY
 
 | | Crystal structure of the native monooxygenase domain of MICAL at 1.45 A resolution | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Siebold, C, Berrow, N, Walter, T.S, Harlos, K, Owens, R.J, Terman, J.R, Stuart, D.I, Kolodkin, A.L, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2005-05-13 | | Release date: | 2005-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-Resolution Structure of the Catalytic Region of Mical (Molecule Interacting with Casl), a Multidomain Flavoenzyme-Signaling Molecule.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2ZEV
 
 | | S. Cerevisiae Geranylgeranyl Pyrophosphate Synthase in Complex with Magnesium, IPP and BPH-715 | | Descriptor: | 3-(DECYLOXY)-1-(2,2-DIPHOSPHONOETHYL)PYRIDINIUM, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, Geranylgeranyl pyrophosphate synthetase, ... | | Authors: | Guo, R.T, Chen, C.K.-M, Cao, R, Oldfield, E, Wang, A.H.-J. | | Deposit date: | 2007-12-17 | | Release date: | 2008-12-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Lipophilic bisphosphonates as dual farnesyl/geranylgeranyl diphosphate synthase inhibitors: an X-ray and NMR investigation
J.Am.Chem.Soc., 131, 2009
|
|
3WQ0
 
 | |
1XG2
 
 | | Crystal structure of the complex between pectin methylesterase and its inhibitor protein | | Descriptor: | Pectinesterase 1, Pectinesterase inhibitor | | Authors: | Di Matteo, A, Raiola, A, Camardella, L, Giovane, A, Bonivento, D, De Lorenzo, G, Cervone, F, Bellincampi, D, Tsernoglou, D. | | Deposit date: | 2004-09-16 | | Release date: | 2005-03-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Interaction between Pectin Methylesterase and a Specific Inhibitor Protein
Plant Cell, 17, 2005
|
|
2H1B
 
 | | ResA E80Q | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Thiol-disulfide oxidoreductase resA | | Authors: | Lewin, A, Crow, A, Oubrie, A, Le Brun, N.E. | | Deposit date: | 2006-05-16 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Molecular Basis for Specificity of the Extracytoplasmic Thioredoxin ResA.
J.Biol.Chem., 281, 2006
|
|
6M4W
 
 | | Crystal structure of MBP fused split FKBP-FRB T2098L mutant in complex with rapamycin | | Descriptor: | GLYCEROL, Peptidyl-prolyl cis-trans isomerase FKBP1A, RAPAMYCIN IMMUNOSUPPRESSANT DRUG, ... | | Authors: | Kikuchi, M, Wu, D, Inoue, T, Umehara, T. | | Deposit date: | 2020-03-09 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Rational design and implementation of a chemically inducible heterotrimerization system.
Nat.Methods, 17, 2020
|
|
3QAK
 
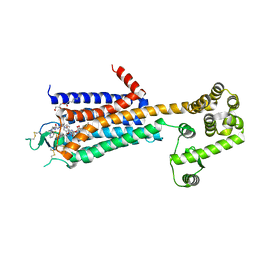 | | Agonist bound structure of the human adenosine A2a receptor | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 6-(2,2-diphenylethylamino)-9-[(2R,3R,4S,5S)-5-(ethylcarbamoyl)-3,4-dihydroxy-oxolan-2-yl]-N-[2-[(1-pyridin-2-ylpiperidin-4-yl)carbamoylamino]ethyl]purine-2-carboxamide, Adenosine receptor A2a,lysozyme chimera | | Authors: | Xu, F, Wu, H, Katritch, V, Han, G.W, Cherezov, V, Stevens, R, GPCR Network (GPCR) | | Deposit date: | 2011-01-11 | | Release date: | 2011-03-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structure of an agonist-bound human A2A adenosine receptor.
Science, 332, 2011
|
|
2YPJ
 
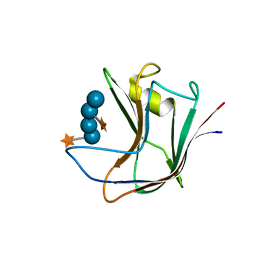 | | Non-catalytic carbohydrate binding module CBM65B | | Descriptor: | ENDOGLUCANASE CEL5A, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Luis, A.S, Venditto, I, Prates, J.A.M, Ferreira, L.M.A, Temple, M.J, Rogowski, A, Basle, A, Xue, J, Knox, J.P, Gilbert, H.J, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2012-10-30 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Understanding How Non-Catalytic Carbohydrate Binding Modules Can Display Specificity for Xyloglucan
J.Biol.Chem., 288, 2013
|
|
2G84
 
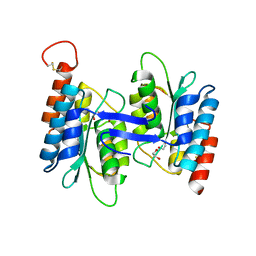 | | Cytidine and deoxycytidylate deaminase zinc-binding region from Nitrosomonas europaea. | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, Cytidine and deoxycytidylate deaminase zinc-binding region, ... | | Authors: | Osipiuk, J, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-01 | | Release date: | 2006-04-04 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray crystal structure of Cytidine and deoxycytidylate deaminase zinc-binding region from Nitrosomonas europaea.
To be Published
|
|
2OQL
 
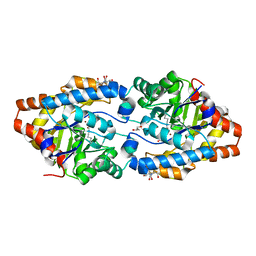 | | Structure of Phosphotriesterase mutant H254Q/H257F | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Parathion hydrolase, ... | | Authors: | Kim, J, Tsai, P, Raushel, F.M, Almo, S.C. | | Deposit date: | 2007-01-31 | | Release date: | 2008-02-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of PTE mutant H254Q/H257F
To be Published
|
|
4OJE
 
 | |
2LD2
 
 | |
4KPE
 
 | | Novel fluoroquinolones in complex with topoisomerase IV from S. pneumoniae and E-site G-gate | | Descriptor: | (7aR,8R)-8-amino-4-cyclopropyl-12-fluoro-1-oxo-4,7,7a,8,9,10-hexahydro-1H-pyrrolo[1',2':1,7]azepino[2,3-h]quinoline-2-carboxylic acid, DNA topoisomerase 4 subunit A, DNA topoisomerase 4 subunit B, ... | | Authors: | Laponogov, I, Pan, X.-S, Vesekov, D.A, Cirz, R.T, Wagman, A.S, Moser, H.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2013-05-13 | | Release date: | 2014-11-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | Exploring the active site of the Streptococcus pneumoniae topoisomerase IV-DNA cleavage complex with novel 7,8-bridged fluoroquinolones.
Open Biol, 6, 2016
|
|
5Q1T
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCLRE1A in complex with FMOPL000492a | | Descriptor: | 1-(phenylmethyl)-4-pyrrol-1-yl-piperidine, DNA cross-link repair 1A protein, MALONATE ION, ... | | Authors: | Newman, J.A, Aitkenhead, H, Lee, S.Y, Kupinska, K, Burgess-Brown, N, Tallon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-15 | | Release date: | 2018-08-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
4KS2
 
 | | Influenza Neuraminidase in complex with antiviral compound (3S,4R,5R)-4-(acetylamino)-3-carbamimidamido-5-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid | | Descriptor: | (3S,4R,5R)-4-(acetylamino)-3-carbamimidamido-5-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, CALCIUM ION, Neuraminidase | | Authors: | Kerry, P.S, Russell, R.J.M. | | Deposit date: | 2013-05-17 | | Release date: | 2013-10-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Structural basis for a class of nanomolar influenza A neuraminidase inhibitors.
Sci Rep, 3, 2013
|
|
1DP0
 
 | | E. COLI BETA-GALACTOSIDASE AT 1.7 ANGSTROM | | Descriptor: | BETA-GALACTOSIDASE, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Juers, D.H, Jacobson, R.H, Wigley, D, Zhang, X.J, Huber, R.E, Tronrud, D.E, Matthews, B.W. | | Deposit date: | 1999-12-22 | | Release date: | 2001-02-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High resolution refinement of beta-galactosidase in a new crystal form reveals multiple metal-binding sites and provides a structural basis for alpha-complementation.
Protein Sci., 9, 2000
|
|
6C6C
 
 | | Structure of glycolipid aGSA[20,6P] in complex with mouse CD1d | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, Beta-2-microglobulin, ... | | Authors: | Zajonc, D.M, Wang, J. | | Deposit date: | 2018-01-18 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A molecular switch in mouse CD1d modulates natural killer T cell activation by alpha-galactosylsphingamides.
J.Biol.Chem., 294, 2019
|
|
2OXB
 
 | | Crystal structure of a cell-wall invertase (E203Q) from Arabidopsis thaliana in complex with sucrose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-fructofuranosidase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Lammens, W, Le Roy, K, Van Laere, A, Van den Ende, W, Rabijns, A. | | Deposit date: | 2007-02-20 | | Release date: | 2008-01-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An alternate sucrose binding mode in the E203Q Arabidopsis invertase mutant: An X-ray crystallography and docking study.
Proteins, 71, 2007
|
|
1QQK
 
 | | THE CRYSTAL STRUCTURE OF FIBROBLAST GROWTH FACTOR 7 (KERATINOCYTE GROWTH FACTOR) | | Descriptor: | FIBROBLAST GROWTH FACTOR 7 | | Authors: | Ye, S, Luo, Y, Pelletier, H, McKeehan, W.L. | | Deposit date: | 1999-06-07 | | Release date: | 2000-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for interaction of FGF-1, FGF-2, and FGF-7 with different heparan sulfate motifs.
Biochemistry, 40, 2001
|
|
2B0C
 
 | | The crystal structure of the putative phosphatase from Escherichia coli | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, MAGNESIUM ION, putative phosphatase | | Authors: | Zhang, R, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-09-13 | | Release date: | 2005-11-22 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0A crystal structure of the putative phosphatase from Escherichia coli
To be Published
|
|
6K9T
 
 | | Crystal structure of a class C beta-lactamase in complex with cefotaxime | | Descriptor: | Beta-lactamase, CEFOTAXIME, C3' cleaved, ... | | Authors: | Bae, D.W, Jung, Y.E, An, Y.J, Na, J.H, Cha, S.S. | | Deposit date: | 2019-06-17 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.459 Å) | | Cite: | Structural Insights into Catalytic Relevances of Substrate Poses in ACC-1.
Antimicrob.Agents Chemother., 63, 2019
|
|
1OCQ
 
 | | COMPLEX OF THE ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHEARANS AT 1.08 ANGSTROM RESOLUTION with cellobio-derived isofagomine | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, ENDOGLUCANASE 5A, GLYCEROL, ... | | Authors: | Varrot, A, Macdonald, J, Stick, R.V, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-02-09 | | Release date: | 2003-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Direct Observation of the Protonation State of an Imino Sugar Glycosidase Inhibitor Upon Binding
J.Am.Chem.Soc., 125, 2003
|
|
2KFX
 
 | |
