7EQD
 
 | | STRUCTURE OF PHOTOSYNTHETIC LH1-RC SUPER-COMPLEX OF RHODOSPIRILLUM RUBRUM | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 2-azanyl-5-[(2~{E},6~{E},8~{E},10~{E},12~{E},14~{E},18~{E},22~{E},26~{E},30~{E},34~{E})-3,7,11,15,19,23,27,31,35,39-decamethyltetraconta-2,6,8,10,12,14,18,22,26,30,34,38-dodecaenyl]-3-methoxy-6-methyl-cyclohexa-2,5-diene-1,4-dione, CARDIOLIPIN, ... | | Authors: | Tani, K, Kanno, R, Ji, X.-C, Yu, L.-J, Hall, M, Kimura, Y, Madigan, M.T, Mizoguchi, A, Humbel, B.M, Wang-Otomo, Z.-Y. | | Deposit date: | 2021-05-01 | | Release date: | 2021-08-18 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Cryo-EM Structure of the Photosynthetic LH1-RC Complex from Rhodospirillum rubrum .
Biochemistry, 2021
|
|
4B9K
 
 | | pVHL-ELOB-ELOC complex_(2S,4R)-1-(3-amino-2-methylbenzoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide bound | | Descriptor: | (2S,4R)-1-(3-amino-2-methylbenzoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide, ACETATE ION, TRANSCRIPTION ELONGATION FACTOR B POLYPEPTIDE 1, ... | | Authors: | Buckley, D.L, Gustafson, J.L, VanMolle, I, Roth, A.G, SeopTae, H, Gareiss, P.C, Jorgensen, W.L, Ciulli, A, Crews, C.M. | | Deposit date: | 2012-09-05 | | Release date: | 2012-10-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small-Molecule Inhibitors of the Interaction between the E3 Ligase Vhl and Hif1 Alpha
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
2G8L
 
 | |
2NVA
 
 | | The X-ray crystal structure of the Paramecium bursaria Chlorella virus arginine decarboxylase bound to agmatine | | Descriptor: | (4-{[(4-{[AMINO(IMINO)METHYL]AMINO}BUTYL)AMINO]METHYL}-5-HYDROXY-6-METHYLPYRIDIN-3-YL)METHYL DIHYDROGEN PHOSPHATE, arginine decarboxylase, A207R protein | | Authors: | Shah, R.H, Akella, R, Goldsmith, E, Phillips, M.A. | | Deposit date: | 2006-11-11 | | Release date: | 2007-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray Structure of Paramecium bursaria Chlorella Virus Arginine Decarboxylase: Insight into the Structural Basis for Substrate Specificity.
Biochemistry, 46, 2007
|
|
2BV7
 
 | | Crystal structure of GLTP with bound GM3 | | Descriptor: | GLYCOLIPID TRANSFER PROTEIN, N-{1-[(HEXOPYRANOSYLOXY)METHYL]-2-HYDROXYNONADECYL}TETRACOSANAMIDE, SULFATE ION | | Authors: | Kidron, H, Airenne, T.T, Nymalm, Y, Nylund, M, West, G, Mattjus, P, Salminen, T.A. | | Deposit date: | 2005-06-23 | | Release date: | 2005-12-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Evidence for Adaptive Ligand Binding of Glycolipid Transfer Protein.
J.Mol.Biol., 355, 2006
|
|
6EPK
 
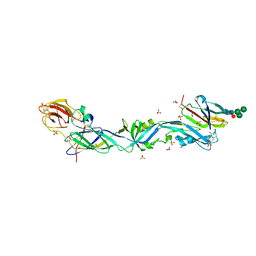 | | CRYSTAL STRUCTURE OF THE PRECURSOR MEMBRANE PROTEIN-ENVELOPE PROTEIN HETERODIMER FROM THE YELLOW FEVER VIRUS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, GLYCEROL, ... | | Authors: | Rey, F.A, Duquerroy, S, Crampon, E, Barba-Spaeth, G. | | Deposit date: | 2017-10-11 | | Release date: | 2018-10-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | New insight into flavivirus maturation from structure/function studies of the yellow fever virus envelope protein complex
Mbio, 2023
|
|
3R6V
 
 | | Crystal structure of aspartase from Bacillus sp. YM55-1 with bound L-aspartate | | Descriptor: | ASPARTIC ACID, Aspartase, CALCIUM ION | | Authors: | Fibriansah, G, Puthan Veetil, V, Poelarends, G.J, Thunnissen, A.-M.W.H. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the catalytic mechanism of aspartate ammonia lyase.
Biochemistry, 50, 2011
|
|
2GGE
 
 | | Crystal Structure of Mandelate Racemase/Muconate Lactonizing Enzyme from Bacillus Subtilis complexed with MG++ at 1.8 A | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, yitF | | Authors: | Malashkevich, V.N, Sauder, J.M, Schwinn, K.D, Emtage, S, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Burley, S.K, Sali, A, Babbitt, P, Pieper, U, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-03-23 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of Mandelate Racemase/Muconate Lactonizing Enzyme from Bacillus Subtilis complexed with MG++ at 1.8 A
To be Published
|
|
2O4T
 
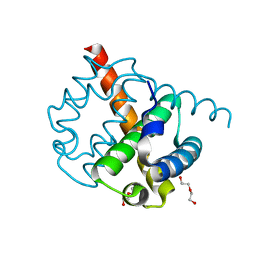 | |
11GS
 
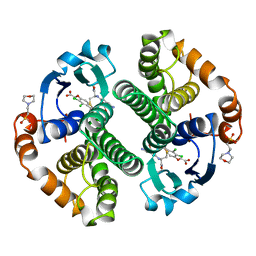 | | Glutathione s-transferase complexed with ethacrynic acid-glutathione conjugate (form ii) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ETHACRYNIC ACID, GLUTATHIONE, ... | | Authors: | Oakley, A.J, Lo Bello, M, Mazzetti, A.P, Federici, G, Parker, M.W. | | Deposit date: | 1997-11-03 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The glutathione conjugate of ethacrynic acid can bind to human pi class glutathione transferase P1-1 in two different modes.
FEBS Lett., 419, 1997
|
|
4JT5
 
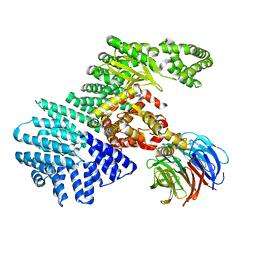 | | mTORdeltaN-mLST8-pp242 complex | | Descriptor: | 2-[4-amino-1-(propan-2-yl)-1H-pyrazolo[3,4-d]pyrimidin-3-yl]-1H-indol-5-ol, Serine/threonine-protein kinase mTOR, Target of rapamycin complex subunit LST8 | | Authors: | Pavletich, N.P, Yang, H. | | Deposit date: | 2013-03-22 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | mTOR kinase structure, mechanism and regulation.
Nature, 497, 2013
|
|
3BGX
 
 | | E. coli Thymidylate Synthase C146S mutant complexed with dTMP and MTF | | Descriptor: | N-({4-[(6aR)-3-amino-1-oxo-1,2,5,6,6a,7-hexahydroimidazo[1,5-f]pteridin-8(9H)-yl]phenyl}carbonyl)-L-glutamic acid, SULFATE ION, THYMIDINE-5'-PHOSPHATE, ... | | Authors: | Roberts, S.A, Montfort, W.R. | | Deposit date: | 2007-11-27 | | Release date: | 2008-12-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Studies of Early Events in Catalysis by Thymidylate Synthase
To be Published
|
|
7GG9
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ANT-DIA-62e4526e-1 (Mpro-x12204) | | Descriptor: | 2-(1H-1,2,3-benzotriazol-1-yl)-1-(4-methylpiperidin-1-yl)ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
4JXF
 
 | | Crystal Structure of PLK4 Kinase with an inhibitor: 400631 ((1R,2S)-2-{3-[(E)-2-{4-[(DIMETHYLAMINO)METHYL]PHENYL}ETHENYL]-2H-INDAZOL-6-YL}-5'-METHOXYSPIRO[CYCLOPROPANE-1,3'-INDOL]-2'(1'H)-ONE) | | Descriptor: | (1R,2S)-2-{3-[(E)-2-{4-[(dimethylamino)methyl]phenyl}ethenyl]-2H-indazol-6-yl}-5'-methoxyspiro[cyclopropane-1,3'-indol]-2'(1'H)-one, 1,2-ETHANEDIOL, Serine/threonine-protein kinase PLK4 | | Authors: | Qiu, W, Plotnikova, O, Feher, M, Awrey, D.E, Chirgadze, N.Y. | | Deposit date: | 2013-03-28 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of PLK4 Kinase with an inhibitor: 400631
To be Published
|
|
3ZZJ
 
 | | Structure of an engineered aspartate aminotransferase | | Descriptor: | ASPARTATE AMINOTRANSFERASE, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fernandez, F.J, deVries, D, Pena-Soler, E, Coll, M, Christen, P, Gehring, H, Vega, M.C. | | Deposit date: | 2011-09-01 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Mechanism of a Cysteine Sulfinate Desulfinase Engineered on the Aspartate Aminotransferase Scaffold.
Biocim.Biophys.Acta, 1824, 2011
|
|
2LPE
 
 | |
5E7G
 
 | |
5DKJ
 
 | | Yeast 20S proteasome in complex with octreotide-PI | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Beck, P, Cui, H, Groll, M. | | Deposit date: | 2015-09-03 | | Release date: | 2015-10-28 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Targeted Delivery of Proteasome Inhibitors to Somatostatin-Receptor-Expressing Cancer Cells by Octreotide Conjugation.
Chemmedchem, 10, 2015
|
|
2LE3
 
 | |
3ZVU
 
 | | Structure of the PYR1 His60Pro mutant in complex with the HAB1 phosphatase and Abscisic acid | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, ABSCISIC ACID RECEPTOR PYR1, MANGANESE (II) ION, ... | | Authors: | Betz, K, Dupeux, F, Santiago, J, Rodriguez, P.L, Marquez, J.A. | | Deposit date: | 2011-07-27 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Thermodynamic Switch Modulates Abscisic Acid Receptor Sensitivity.
Embo J., 30, 2011
|
|
6EZN
 
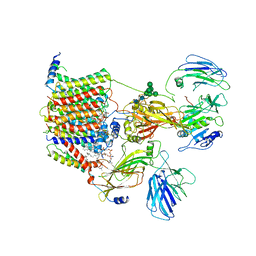 | | Cryo-EM structure of the yeast oligosaccharyltransferase (OST) complex | | Descriptor: | 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wild, R, Kowal, J, Eyring, J, Ngwa, E.M, Aebi, M, Locher, K.P. | | Deposit date: | 2017-11-16 | | Release date: | 2018-01-17 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the yeast oligosaccharyltransferase complex gives insight into eukaryotic N-glycosylation.
Science, 359, 2018
|
|
4RUP
 
 | | Crystal structure of zVDR L337H mutant-Gemini72 complex | | Descriptor: | (1R,3R,7E,17beta)-17-[(1R)-6,6,6-trifluoro-5-hydroxy-1-(4-hydroxy-4-methylpentyl)-5-(trifluoromethyl)hex-3-yn-1-yl]-9,1 0-secoestra-5,7-diene-1,3-diol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Huet, T, Moras, D, Rochel, N. | | Deposit date: | 2014-11-21 | | Release date: | 2015-10-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A vitamin D receptor selectively activated by gemini analogs reveals ligand dependent and independent effects.
Cell Rep, 10, 2015
|
|
1RDP
 
 | | Cholera Toxin B-Pentamer Complexed With Bivalent Nitrophenol-Galactoside Ligand BV3 | | Descriptor: | 1,3-BIS-([[3-(4-{3-[3-NITRO-5-(GALACTOPYRANOSYLOXY)-BENZOYLAMINO]-PROPYL}-PIPERAZIN-1-YL)-PROPYLAMINO-3,4-DIOXO-CYCLOBU TENYL]-AMINO-ETHYL]-AMINO-CARBONYLOXY)-2-AMINO-PROPANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, TRIETHYLENE GLYCOL, ... | | Authors: | Pickens, J.C, Mitchell, D.D, Liu, J, Tan, X, Zhang, Z, Verlinde, C.L, Hol, W.G, Fan, E. | | Deposit date: | 2003-11-05 | | Release date: | 2004-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Nonspanning bivalent ligands as improved surface receptor binding inhibitors of the cholera toxin B pentamer.
Chem.Biol., 11, 2004
|
|
2LA7
 
 | | NMR structure of the protein YP_557733.1 from Burkholderia xenovorans | | Descriptor: | Uncharacterized protein | | Authors: | Jaudzems, K, Serrano, P, Michael, G, Reto, H, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2011-03-04 | | Release date: | 2011-03-30 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the protein YP_557733.1 from Burkholderia xenovorans
To be Published
|
|
4JFP
 
 | | A2 HLA complex with G4A heteroclitic variant of Melanoma peptide | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-2-microglobulin, ... | | Authors: | Rizkallah, P.J, Cole, D.K, Madura, F, Sewell, A.K. | | Deposit date: | 2013-02-28 | | Release date: | 2013-05-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | T-cell receptor specificity maintained by altered thermodynamics.
J.Biol.Chem., 288, 2013
|
|
