2RGM
 
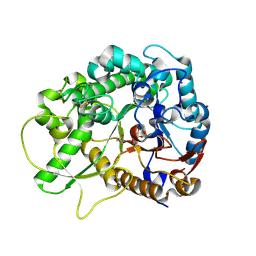 | | Rice BGlu1 beta-glucosidase, a plant exoglucanase/beta-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-deoxy-2-fluoro-alpha-D-glucopyranose, Beta-glucosidase, ... | | Authors: | Chuenchor, W, Ketudat Cairns, J.R, Pengthaisong, S, Robinson, R.C, Yuvaniyama, J, Chen, C.-J. | | Deposit date: | 2007-10-04 | | Release date: | 2008-02-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Insights into Rice BGlu1 beta-Glucosidase Oligosaccharide Hydrolysis and Transglycosylation
J.Mol.Biol., 377, 2008
|
|
2E2N
 
 | | Crystal structure of Sulfolobus tokodaii hexokinase in the apo form | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, HEXOKINASE, SULFATE ION | | Authors: | Nishimasu, H, Fushinobu, S, Shoun, H, Wakagi, T. | | Deposit date: | 2006-11-15 | | Release date: | 2007-01-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of an ATP-dependent hexokinase with broad substrate specificity from the hyperthermophilic archaeon Sulfolobus tokodaii.
J.Biol.Chem., 282, 2007
|
|
1DMY
 
 | |
1DZ6
 
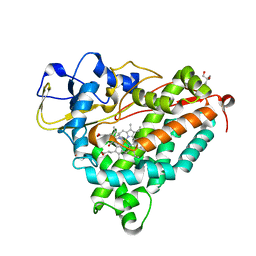 | | ferrous p450cam from pseudomonas putida | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, CYTOCHROME P450-CAM, ... | | Authors: | Schlichting, I, Berendzen, J, Chu, K, Stock, A.M, Maves, S.A, Benson, D.E, Sweet, R.M, Ringe, D, Petsko, G.A, Sligar, S.G. | | Deposit date: | 2000-02-18 | | Release date: | 2000-03-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Catalytic Pathway of Cytochrome P450Cam at Atomic Resolution
Science, 287, 2000
|
|
1DZ8
 
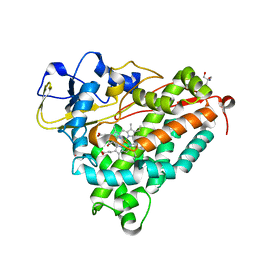 | | oxygen complex of p450cam from pseudomonas putida | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, CYTOCHROME P450-CAM, ... | | Authors: | Schlichting, I, Berendzen, J, Chu, K, Stock, A.M, Maves, S.A, Benson, D.E, Sweet, R.M, Ringe, D, Petsko, G.A, Sligar, S.G. | | Deposit date: | 2000-02-18 | | Release date: | 2000-03-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Catalytic Pathway of Cytochrome P450Cam at Atomic Resolution
Science, 287, 2000
|
|
2HGX
 
 | |
2HHF
 
 | |
1E5E
 
 | | METHIONINE GAMMA-LYASE (MGL) FROM TRICHOMONAS VAGINALIS IN COMPLEX WITH PROPARGYLGLYCINE | | Descriptor: | GLYCEROL, METHIONINE GAMMA-LYASE, N-(HYDROXY{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)NORVALINE, ... | | Authors: | Goodall, G, Mottram, J.C, Coombs, G.H, Lapthorn, A.J. | | Deposit date: | 2000-07-25 | | Release date: | 2001-07-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | The Structure and Proposed Catalytic Mechanism of Methionine Gamma-Lyase
To be Published
|
|
1EGW
 
 | | CRYSTAL STRUCTURE OF MEF2A CORE BOUND TO DNA | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*CP*TP*AP*TP*TP*AP*TP*TP*AP*GP*CP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*GP*CP*TP*AP*AP*TP*AP*AP*TP*AP*GP*CP*TP*T)-3'), MADS BOX TRANSCRIPTION ENHANCER FACTOR 2, ... | | Authors: | Santelli, E, Richmond, T.J. | | Deposit date: | 2000-02-17 | | Release date: | 2000-03-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of MEF2A core bound to DNA at 1.5 A resolution.
J.Mol.Biol., 297, 2000
|
|
1EHX
 
 | | NMR SOLUTION STRUCTURE OF THE LAST UNKNOWN MODULE OF THE CELLULOSOMAL SCAFFOLDIN PROTEIN CIPC OF CLOSTRIDUM CELLULOLYTICUM | | Descriptor: | SCAFFOLDIN PROTEIN | | Authors: | Mosbah, A, Belaich, A, Bornet, O, Belaich, J.P, Henrissat, B, Darbon, H. | | Deposit date: | 2000-02-23 | | Release date: | 2000-11-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the module X2 1 of unknown function of the cellulosomal scaffolding protein CipC of Clostridium cellulolyticum.
J.Mol.Biol., 304, 2000
|
|
2HGW
 
 | |
1ECA
 
 | |
2MCT
 
 | | NMR structure of the protein ZP_02042476.1 from Ruminococcus gnavus | | Descriptor: | Uncharacterized protein | | Authors: | Martin, B.T, Serrano, P, Geralt, M, Dutta, S, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2013-08-27 | | Release date: | 2013-11-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the protein ZP_02042476.1 from Ruminococcus gnavus.
To be Published
|
|
2HH0
 
 | |
2MCA
 
 | | NMR structure of the protein YP_002937094.1 from Eubacterium rectale | | Descriptor: | Uncharacterized protein | | Authors: | Proudfoot, A, Serrano, P, Geralt, M, Dutta, S, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2013-08-19 | | Release date: | 2013-09-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the protein YP_002937094.1 from Eubacterium rectale
To be Published
|
|
2MVB
 
 | |
2MQB
 
 | |
2MWM
 
 | |
3WHU
 
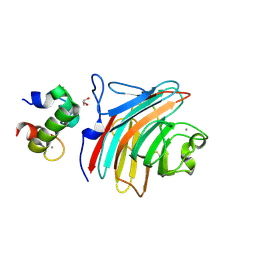 | | Crystal structure of ERGIC-53/MCFD2, Calcium/Man2-bound form | | Descriptor: | CALCIUM ION, GLYCEROL, Multiple coagulation factor deficiency protein 2, ... | | Authors: | Satoh, T, Suzuki, K, Kato, K. | | Deposit date: | 2013-08-30 | | Release date: | 2014-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Disparate Sugar-Binding Specificities in the Homologous Cargo Receptors ERGIC-53 and VIP36
Plos One, 9, 2014
|
|
1UEP
 
 | | Solution Structure of The Third PDZ Domain of Human Atrophin-1 Interacting Protein 1 (KIAA0705 Protein) | | Descriptor: | Membrane Associated Guanylate Kinase Inverted-2 (MAGI-2) | | Authors: | Miyamoto, K, Kigawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-20 | | Release date: | 2003-11-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of The Third PDZ Domain of Human Atrophin-1 Interacting Protein 1 (KIAA0705 Protein)
To be Published
|
|
2YFE
 
 | |
4MGD
 
 | | Crystal structure of hERa-LBD (Y537S) in complex with HPTE | | Descriptor: | 1,2-ETHANEDIOL, 4,4'-(2,2,2-trichloroethane-1,1-diyl)diphenol, Estrogen receptor, ... | | Authors: | Delfosse, V, Grimaldi, M, Bourguet, W. | | Deposit date: | 2013-08-28 | | Release date: | 2014-09-03 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional profiling of environmental ligands for estrogen receptors.
Environ.Health Perspect., 122, 2014
|
|
3NZ2
 
 | | Crystal Structure of Hexapeptide-Repeat containing-Acetyltransferase VCA0836 Complexed with Acetyl Co Enzyme A from Vibrio cholerae O1 biovar eltor | | Descriptor: | 1,4-BUTANEDIOL, ACETIC ACID, ACETYL COENZYME *A, ... | | Authors: | Kim, Y, Maltseva, N, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-15 | | Release date: | 2010-08-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Hexapeptide-Repeat containing-Acetyltransferase VCA0836 Complexed with Acetyl Co Enzyme A from Vibrio cholerae O1 biovar eltor
To be Published
|
|
3WQ1
 
 | |
3O28
 
 | | Ligand-binding domain of GluA2 (flip) ionotropic glutamate receptor in complex with an allosteric modulator | | Descriptor: | 2-({[3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl]acetyl}amino)-4,5,6,7-tetrahydro-1-benzothiophene-3-carboxamide, DIMETHYL SULFOXIDE, GLUTAMIC ACID, ... | | Authors: | Maclean, J.K.F, Basten, S, Campbell, R.A, Cumming, I.A, Gillen, K.J, Gillespie, J, Jamieson, C, Kazemier, B, Kiczun, M, Lamont, Y, Lyons, A.J, Moir, E.M, Morrow, J.A, Papakosta, M, Rankovic, Z, Smith, L. | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-15 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel series of positive modulators of the AMPA receptor: discovery and structure based hit-to-lead studies.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
